Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy
- PMID: 22505714
- PMCID: PMC3365942
- DOI: 10.1074/jbc.M111.322933
Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy
Abstract
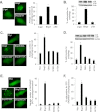
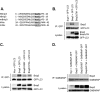
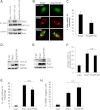
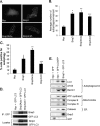
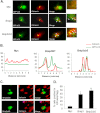
Autophagy plays an important role in cellular quality control and is responsible for removing protein aggregates and dysfunctional organelles. Bnip3 is an atypical BH3-only protein that is known to cause mitochondrial dysfunction and cell death. Interestingly, Bnip3 can also protect against cell death by inducing mitochondrial autophagy. The mechanism for this process, however, remains poorly understood. Bnip3 contains a C-terminal transmembrane domain that is essential for homodimerization and proapoptotic function. In this study, we show that homodimerization of Bnip3 is also a requirement for induction of autophagy. Several Bnip3 mutants that do not interfere with its mitochondrial localization but disrupt homodimerization failed to induce autophagy in cells. In addition, we discovered that endogenous Bnip3 is localized to both mitochondria and the endoplasmic reticulum (ER). To investigate the effects of Bnip3 at mitochondria or the ER on autophagy, Bnip3 was targeted specifically to each organelle by substituting the Bnip3 transmembrane domain with that of Acta or cytochrome b(5). We found that Bnip3 enhanced autophagy in cells from both sites. We also discovered that Bnip3 induced removal of both ER (ERphagy) and mitochondria (mitophagy) via autophagy. The clearance of these organelles was mediated in part via binding of Bnip3 to LC3 on the autophagosome. Although ablation of the Bnip3-LC3 interaction by mutating the LC3 binding site did not impair the prodeath activity of Bnip3, it significantly reduced both mitophagy and ERphagy. Our data indicate that Bnip3 regulates the apoptotic balance as an autophagy receptor that induces removal of both mitochondria and ER.
Figures
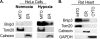

Similar articles
-
Mitophagy regulates mitochondrial network signaling, oxidative stress, and apoptosis during myoblast differentiation.Autophagy. 2019 Sep;15(9):1606-1619. doi: 10.1080/15548627.2019.1591672. Epub 2019 Apr 7. Autophagy. 2019. PMID: 30859901 Free PMC article.
-
Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore.Autophagy. 2010 Oct;6(7):855-62. doi: 10.4161/auto.6.7.13005. Autophagy. 2010. PMID: 20668412 Free PMC article.
-
BNIP3 mediates cell death by different pathways following localization to endoplasmic reticulum and mitochondrion.FASEB J. 2009 Oct;23(10):3405-14. doi: 10.1096/fj.08-124354. Epub 2009 Jun 17. FASEB J. 2009. PMID: 19535684 Free PMC article.
-
Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B interacting protein 3 and Nip-like protein X.Antioxid Redox Signal. 2011 May 15;14(10):1959-69. doi: 10.1089/ars.2010.3772. Epub 2011 Mar 4. Antioxid Redox Signal. 2011. PMID: 21126215 Free PMC article. Review.
-
[BNIP3 as an atypical representative of the Bcl-2 protein family. Part 1: BNIP3, a regulator of non-apoptotic programmed cell death].Postepy Hig Med Dosw (Online). 2009 Sep 10;63:409-17. Postepy Hig Med Dosw (Online). 2009. PMID: 19745227 Review. Polish.
Cited by
-
Endoplasmic Reticulum (ER) and ER-Phagy.Prog Mol Subcell Biol. 2021;59:99-114. doi: 10.1007/978-3-030-67696-4_5. Prog Mol Subcell Biol. 2021. PMID: 34050863
-
Long-term HIF-1α stabilization reduces respiration, promotes mitophagy, and results in retinal cell death.Sci Rep. 2023 Nov 23;13(1):20541. doi: 10.1038/s41598-023-47942-8. Sci Rep. 2023. PMID: 37996657 Free PMC article.
-
A PPARγ-Bnip3 Axis Couples Adipose Mitochondrial Fusion-Fission Balance to Systemic Insulin Sensitivity.Diabetes. 2016 Sep;65(9):2591-605. doi: 10.2337/db16-0243. Epub 2016 Jun 20. Diabetes. 2016. PMID: 27325287 Free PMC article.
-
Hypoxia and Selective Autophagy in Cancer Development and Therapy.Front Cell Dev Biol. 2018 Sep 10;6:104. doi: 10.3389/fcell.2018.00104. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30250843 Free PMC article. Review.
-
Increased autophagy in EphrinB2-deficient osteocytes is associated with elevated secondary mineralization and brittle bone.Nat Commun. 2019 Jul 31;10(1):3436. doi: 10.1038/s41467-019-11373-9. Nat Commun. 2019. PMID: 31366886 Free PMC article.
References
-
- Levine B., Klionsky D. J. (2004) Development by self-digestion. Molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 - PubMed
-
- Hamasaki M., Noda T., Baba M., Ohsumi Y. (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65 - PubMed
-
- Bernales S., Schuck S., Walter P. (2007) ER-phagy. Selective autophagy of the endoplasmic reticulum. Autophagy 3, 285–287 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

