SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing
- PMID: 22506599
- PMCID: PMC3342519
- DOI: 10.1089/cmb.2012.0021
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing
Abstract
The lion's share of bacteria in various environments cannot be cloned in the laboratory and thus cannot be sequenced using existing technologies. A major goal of single-cell genomics is to complement gene-centric metagenomic data with whole-genome assemblies of uncultivated organisms. Assembly of single-cell data is challenging because of highly non-uniform read coverage as well as elevated levels of sequencing errors and chimeric reads. We describe SPAdes, a new assembler for both single-cell and standard (multicell) assembly, and demonstrate that it improves on the recently released E+V-SC assembler (specialized for single-cell data) and on popular assemblers Velvet and SoapDeNovo (for multicell data). SPAdes generates single-cell assemblies, providing information about genomes of uncultivatable bacteria that vastly exceeds what may be obtained via traditional metagenomics studies. SPAdes is available online ( http://bioinf.spbau.ru/spades ). It is distributed as open source software.
Figures

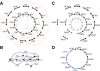



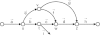
References
-
- Bandeira N. Clauser K. Pevzner P. Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol. Cell Proteomics. 2007;6:1123–1134. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

