Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: carbon and energy flow contribute to the distinct biofilm growth state
- PMID: 22507456
- PMCID: PMC3431258
- DOI: 10.1186/1471-2164-13-138
Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: carbon and energy flow contribute to the distinct biofilm growth state
Abstract
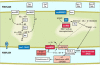
Background: Desulfovibrio vulgaris Hildenborough is a sulfate-reducing bacterium (SRB) that is intensively studied in the context of metal corrosion and heavy-metal bioremediation, and SRB populations are commonly observed in pipe and subsurface environments as surface-associated populations. In order to elucidate physiological changes associated with biofilm growth at both the transcript and protein level, transcriptomic and proteomic analyses were done on mature biofilm cells and compared to both batch and reactor planktonic populations. The biofilms were cultivated with lactate and sulfate in a continuously fed biofilm reactor, and compared to both batch and reactor planktonic populations.
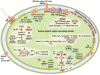
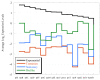
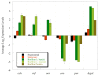
Results: The functional genomic analysis demonstrated that biofilm cells were different compared to planktonic cells, and the majority of altered abundances for genes and proteins were annotated as hypothetical (unknown function), energy conservation, amino acid metabolism, and signal transduction. Genes and proteins that showed similar trends in detected levels were particularly involved in energy conservation such as increases in an annotated ech hydrogenase, formate dehydrogenase, pyruvate:ferredoxin oxidoreductase, and rnf oxidoreductase, and the biofilm cells had elevated formate dehydrogenase activity. Several other hydrogenases and formate dehydrogenases also showed an increased protein level, while decreased transcript and protein levels were observed for putative coo hydrogenase as well as a lactate permease and hyp hydrogenases for biofilm cells. Genes annotated for amino acid synthesis and nitrogen utilization were also predominant changers within the biofilm state. Ribosomal transcripts and proteins were notably decreased within the biofilm cells compared to exponential-phase cells but were not as low as levels observed in planktonic, stationary-phase cells. Several putative, extracellular proteins (DVU1012, 1545) were also detected in the extracellular fraction from biofilm cells.
Conclusions: Even though both the planktonic and biofilm cells were oxidizing lactate and reducing sulfate, the biofilm cells were physiologically distinct compared to planktonic growth states due to altered abundances of genes/proteins involved in carbon/energy flow and extracellular structures. In addition, average expression values for multiple rRNA transcripts and respiratory activity measurements indicated that biofilm cells were metabolically more similar to exponential-phase cells although biofilm cells are structured differently. The characterization of physiological advantages and constraints of the biofilm growth state for sulfate-reducing bacteria will provide insight into bioremediation applications as well as microbially-induced metal corrosion.
Figures




References
-
- Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. Physical constraints affecting bacterial habitats and activity in unsaturated porous media - a review. Adv Water Resources. 2007;30:1505–1527. doi: 10.1016/j.advwatres.2006.05.025. - DOI
-
- Hamilton WA. Sulfate-reducing bacteria: Physiology determines their environmental impact. Geomicrobiol J. 1998;15:19–28. doi: 10.1080/01490459809378059. - DOI
-
- Reardon CL, Cummings DE, Petzke LM, Kinsall BL, Watson DB, Peyton BM, Geesey GG. Composition and diversity of microbial communities recovered from surrogate minerals incubated in an acidic uranium-contaminated aquifer. Appl Environ Microbiol. 2004;70:6037–6046. doi: 10.1128/AEM.70.10.6037-6046.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

