The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae
- PMID: 22508509
- PMCID: PMC3328390
- DOI: 10.1083/jcb.201111115
The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae
Abstract
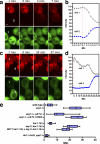
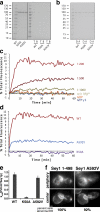
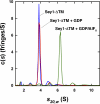
The endoplasmic reticulum (ER) forms a network of tubules and sheets that requires homotypic membrane fusion to be maintained. In metazoans, this process is mediated by dynamin-like guanosine triphosphatases (GTPases) called atlastins (ATLs), which are also required to maintain ER morphology. Previous work suggested that the dynamin-like GTPase Sey1p was needed to maintain ER morphology in Saccharomyces cerevisiae. In this paper, we demonstrate that Sey1p, like ATLs, mediates homotypic ER fusion. The absence of Sey1p resulted in the ER undergoing delayed fusion in vivo and proteoliposomes containing purified Sey1p fused in a GTP-dependent manner in vitro. Sey1p could be partially replaced by ATL1 in vivo. Like ATL1, Sey1p underwent GTP-dependent dimerization. We found that the residual ER-ER fusion that occurred in cells lacking Sey1p required the ER SNARE Ufe1p. Collectively, our results show that Sey1p and its homologues function analogously to ATLs in mediating ER fusion. They also indicate that S. cerevisiae has an alternative fusion mechanism that requires ER SNAREs.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

