Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation
- PMID: 22508689
- PMCID: PMC3382096
- DOI: 10.1096/fj.12-204495
Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation
Abstract
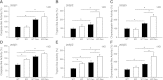
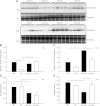
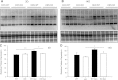
Deletion of muscle RING finger 1 (MuRF1), an E3 ubiquitin ligase, leads to sparing of muscle mass following denervation. The purpose of this study was to test the hypothesis that muscle sparing in mice with a deletion of MuRF1 is due to the selective inhibition of the ubiquitin proteasome system. Activities of the 20S and 26S proteasomes, calpain and cathepsin L, were measured in the triceps surae muscles of wild-type (WT) and MuRF1-knockout (KO) mice at 3 and 14 d following denervation. In addition, fractional protein synthesis rates and differential gene expression were measured in WT and KO muscle. The major finding was that 20S and 26S proteasome activities were significantly elevated (1.5- to 2.5-fold) after 14 d of denervation in both WT and KO mice relative to control, but interestingly, the activities of both the 20S and 26S proteasome were significantly higher in KO than WT mice. Further, mRNA expression of MAFbx was elevated after 14 d of denervation in KO, but not WT, mice. These data challenge the conventional dogma that MuRF1 is controlling the degradation of only contractile proteins and suggest a role for MuRF1 in the global control of the ubiquitin proteasome system and protein turnover.
Figures







References
-
- Taillandier D., Aurousseau E., Meynial-Denis D., Bechet D., Ferrara M., Cottin P., Ducastaing A., Bigard X., Guezennec C. Y., Schmid H. P., Attaix D. (1996) Coordinate activation of lysosomal, Ca2+-activated and ATP-ubiquitin-dependent proteinases in the unweighted rat soleus muscle. Biochem. J. 316, 65–72 - PMC - PubMed
-
- Price S. R., Bailey J. L., Wang X., Jurkovitz C., England B. K., Ding X., Phillips L. S., Mitch W. E. (1996) Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin–proteasome proteolytic pathway by a mechanism including gene transcription. J. Clin. Invest. 98, 1703–1708 - PMC - PubMed
-
- Temparis S., Asensi M., Taillandier D., Aurousseau E., Larbaud D., Obled A., Bechet D., Ferrara M., Estrela J. M., Attaix D. (1994) Increased ATP-ubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Res. 54, 5568–5573 - PubMed
-
- Baracos V. E., DeVivo C., Hoyle D. H., Goldberg A. L. (1995) Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am. J. Physiol. 268, E996–E1006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

