Mutant p53 cooperates with ETS2 to promote etoposide resistance
- PMID: 22508727
- PMCID: PMC3337457
- DOI: 10.1101/gad.181685.111
Mutant p53 cooperates with ETS2 to promote etoposide resistance
Abstract
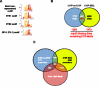
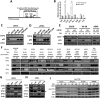
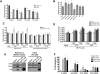
Mutant p53 (mtp53) promotes chemotherapy resistance through multiple mechanisms, including disabling proapoptotic proteins and regulating gene expression. Comparison of genome wide analysis of mtp53 binding revealed that the ETS-binding site motif (EBS) is prevalent within predicted mtp53-binding sites. We demonstrate that mtp53 regulates gene expression through EBS in promoters and that ETS2 mediates the interaction with this motif. Importantly, we identified TDP2, a 5'-tyrosyl DNA phosphodiesterase involved in the repair of DNA damage caused by etoposide, as a transcriptional target of mtp53. We demonstrate that suppression of TDP2 sensitizes mtp53-expressing cells to etoposide and that mtp53 and TDP2 are frequently overexpressed in human lung cancer; thus, our analysis identifies a potentially "druggable" component of mtp53's gain-of-function activity.
Figures




Similar articles
-
Mutant p53 protects ETS2 from non-canonical COP1/DET1 dependent degradation.Oncotarget. 2016 Mar 15;7(11):12554-67. doi: 10.18632/oncotarget.7275. Oncotarget. 2016. PMID: 26871468 Free PMC article.
-
Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing.DNA Repair (Amst). 2016 Jul;43:38-47. doi: 10.1016/j.dnarep.2016.04.009. Epub 2016 May 7. DNA Repair (Amst). 2016. PMID: 27235629
-
WTp53 induction does not override MTp53 chemoresistance and radioresistance due to gain-of-function in lung cancer cells.Mol Cancer Ther. 2008 Apr;7(4):980-92. doi: 10.1158/1535-7163.MCT-07-0471. Mol Cancer Ther. 2008. PMID: 18413811
-
Control of Nucleotide Metabolism Enables Mutant p53's Oncogenic Gain-of-Function Activity.Int J Mol Sci. 2017 Dec 19;18(12):2759. doi: 10.3390/ijms18122759. Int J Mol Sci. 2017. PMID: 29257071 Free PMC article. Review.
-
Mutant p53 and ETS2, a Tale of Reciprocity.Front Oncol. 2016 Feb 18;6:35. doi: 10.3389/fonc.2016.00035. eCollection 2016. Front Oncol. 2016. PMID: 26925389 Free PMC article. Review.
Cited by
-
Mutant p53 Sequestration of the MDM2 Acidic Domain Inhibits E3 Ligase Activity.Mol Cell Biol. 2019 Feb 4;39(4):e00375-18. doi: 10.1128/MCB.00375-18. Print 2019 Feb 15. Mol Cell Biol. 2019. PMID: 30455251 Free PMC article.
-
Prognostic implications and interaction of L1 methylation and p53 expression statuses in advanced gastric cancer.Clin Epigenetics. 2019 May 14;11(1):77. doi: 10.1186/s13148-019-0661-x. Clin Epigenetics. 2019. PMID: 31088544 Free PMC article.
-
High CD49f expression is associated with osteosarcoma tumor progression: a study using patient-derived primary cell cultures.Cancer Med. 2014 Aug;3(4):796-811. doi: 10.1002/cam4.249. Epub 2014 May 7. Cancer Med. 2014. PMID: 24802970 Free PMC article.
-
From regulation to deregulation of p53 in hematologic malignancies: implications for diagnosis, prognosis and therapy.Biomark Res. 2024 Nov 14;12(1):137. doi: 10.1186/s40364-024-00676-9. Biomark Res. 2024. PMID: 39538363 Free PMC article. Review.
-
Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.Nat Struct Mol Biol. 2012 Dec;19(12):1372-7. doi: 10.1038/nsmb.2423. Epub 2012 Oct 28. Nat Struct Mol Biol. 2012. PMID: 23104058 Free PMC article.
References
-
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. 2009. A Mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137: 87–98 - PubMed
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
-
- Basuyaux JP, Ferreira E, Stehelin D, Buttice G 1997. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J Biol Chem 272: 26188–26195 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
