Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters
- PMID: 22508764
- PMCID: PMC3371715
- DOI: 10.1101/gr.136929.111
Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters
Abstract
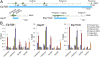
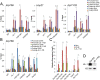
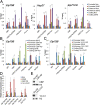
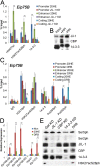
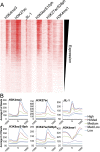
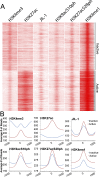
Transcription regulation is mediated by enhancers that bind sequence-specific transcription factors, which in turn interact with the promoters of the genes they control. Here, we show that the JIL-1 kinase is present at both enhancers and promoters of ecdysone-induced Drosophila genes, where it phosphorylates the Ser10 and Ser28 residues of histone H3. JIL-1 is also required for CREB binding protein (CBP)-mediated acetylation of Lys27, a well-characterized mark of active enhancers. The presence of these proteins at enhancers and promoters of ecdysone-induced genes results in the establishment of the H3K9acS10ph and H3K27acS28ph marks at both regulatory sequences. These modifications are necessary for the recruitment of 14-3-3, a scaffolding protein capable of facilitating interactions between two simultaneously bound proteins. Chromatin conformation capture assays indicate that interaction between the enhancer and the promoter is dependent on the presence of JIL-1, 14-3-3, and CBP. Genome-wide analyses extend these conclusions to most Drosophila genes, showing that the presence of JIL-1, H3K9acS10ph, and H3K27acS28ph is a general feature of enhancers and promoters in this organism.
Figures

Similar articles
-
Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila.Genes Dev. 2007 Nov 1;21(21):2818-31. doi: 10.1101/gad.1604007. Epub 2007 Oct 17. Genes Dev. 2007. PMID: 17942706 Free PMC article.
-
Global analysis of the relationship between JIL-1 kinase and transcription.PLoS Genet. 2011 Mar;7(3):e1001327. doi: 10.1371/journal.pgen.1001327. Epub 2011 Mar 10. PLoS Genet. 2011. PMID: 21423663 Free PMC article.
-
Domain requirements of the JIL-1 tandem kinase for histone H3 serine 10 phosphorylation and chromatin remodeling in vivo.J Biol Chem. 2013 Jul 5;288(27):19441-9. doi: 10.1074/jbc.M113.464271. Epub 2013 May 30. J Biol Chem. 2013. PMID: 23723094 Free PMC article.
-
Evaluating Enhancer Function and Transcription.Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
-
Dynamics of histone variant H3.3 and its coregulation with H2A.Z at enhancers and promoters.Nucleus. 2014 Jan-Feb;5(1):21-7. doi: 10.4161/nucl.28067. Epub 2014 Feb 3. Nucleus. 2014. PMID: 24637397 Free PMC article. Review.
Cited by
-
The Drosophila transcription factor Adf-1 (nalyot) regulates dendrite growth by controlling FasII and Staufen expression downstream of CaMKII and neural activity.J Neurosci. 2013 Jul 17;33(29):11916-31. doi: 10.1523/JNEUROSCI.1760-13.2013. J Neurosci. 2013. PMID: 23864680 Free PMC article.
-
The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner.J Cell Sci. 2014 Jul 1;127(Pt 13):2956-66. doi: 10.1242/jcs.151126. Epub 2014 Apr 4. J Cell Sci. 2014. PMID: 24706949 Free PMC article.
-
CREB-binding protein plays key roles in juvenile hormone action in the red flour beetle, Tribolium Castaneum.Sci Rep. 2018 Jan 23;8(1):1426. doi: 10.1038/s41598-018-19667-6. Sci Rep. 2018. PMID: 29362416 Free PMC article.
-
Uncovering the functional constraints underlying the genomic organization of the odorant-binding protein genes.Genome Biol Evol. 2013;5(11):2096-108. doi: 10.1093/gbe/evt158. Genome Biol Evol. 2013. PMID: 24148943 Free PMC article.
-
Transcription factor binding at enhancers: shaping a genomic regulatory landscape in flux.Front Genet. 2012 Sep 28;3:195. doi: 10.3389/fgene.2012.00195. eCollection 2012. Front Genet. 2012. PMID: 23060900 Free PMC article.
References
-
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 - PubMed
-
- Bernardo TJ, Dubrovskaya VA, Jannat H, Maughan B, Dubrovsky EB 2009. Hormonal regulation of the E75 gene in Drosophila: Identifying functional regulatory elements through computational and biological analysis. J Mol Biol 387: 794–808 - PubMed
-
- Cherbas L, Lee K, Cherbas P 1991. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev 5: 120–131 - PubMed
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
