Immunogenicity and cross-reactivity of a representative ancestral sequence in hepatitis C virus infection
- PMID: 22508927
- PMCID: PMC3345099
- DOI: 10.4049/jimmunol.1103008
Immunogenicity and cross-reactivity of a representative ancestral sequence in hepatitis C virus infection
Abstract
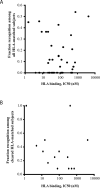
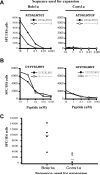
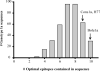
Vaccines designed to prevent or to treat hepatitis C viral infection must achieve maximum cross-reactivity against widely divergent circulating strains. Rational approaches for sequence selection to maximize immunogenicity and minimize genetic distance across circulating strains may enhance vaccine induction of optimal cytotoxic T cell responses. We assessed T cell recognition of potential hepatitis C virus (HCV) vaccine sequences generated using three rational approaches: combining epitopes with predicted tight binding to the MHC, consensus sequence (most common amino acid at each position), and representative ancestral sequence that had been derived using bayesian phylogenetic tools. No correlation was seen between peptide-MHC binding affinity and frequency of recognition, as measured by an IFN-γ T cell response in HLA-matched HCV-infected individuals. Peptides encoding representative, consensus, and natural variant sequences were then tested for the capacity to expand CD8 T cell populations and to elicit cross-reactive CD8 T cell responses. CD8(+) T cells expanded with representative sequence HCV generally more broadly and robustly recognized highly diverse circulating HCV strains than did T cells expanded with either consensus sequence or naturally occurring sequence variants. These data support the use of representative sequence in HCV vaccine design.
Figures


References
-
- Schwartlander B. Global burden of disease. Lancet. 1997;350:141–142. - PubMed
-
- Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 1999;341:556–562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

