Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing
- PMID: 22508932
- PMCID: PMC3375937
- DOI: 10.1104/pp.112.196048
Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing
Abstract
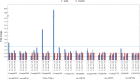
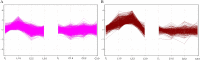
The male sterility of thermosensitive genic male sterile (TGMS) lines of wheat (Triticum aestivum) is strictly controlled by temperature. The early phase of anther development is especially susceptible to cold stress. MicroRNAs (miRNAs) play an important role in plant development and in responses to environmental stress. In this study, deep sequencing of small RNA (smRNA) libraries obtained from spike tissues of the TGMS line under cold and control conditions identified a total of 78 unique miRNA sequences from 30 families and trans-acting small interfering RNAs (tasiRNAs) derived from two TAS3 genes. To identify smRNA targets in the wheat TGMS line, we applied the degradome sequencing method, which globally and directly identifies the remnants of smRNA-directed target cleavage. We identified 26 targets of 16 miRNA families and three targets of tasiRNAs. Comparing smRNA sequencing data sets and TaqMan quantitative polymerase chain reaction results, we identified six miRNAs and one tasiRNA (tasiRNA-ARF [for Auxin-Responsive Factor]) as cold stress-responsive smRNAs in spike tissues of the TGMS line. We also determined the expression profiles of target genes that encode transcription factors in response to cold stress. Interestingly, the expression of cold stress-responsive smRNAs integrated in the auxin-signaling pathway and their target genes was largely noncorrelated. We investigated the tissue-specific expression of smRNAs using a tissue microarray approach. Our data indicated that miR167 and tasiRNA-ARF play roles in regulating the auxin-signaling pathway and possibly in the developmental response to cold stress. These data provide evidence that smRNA regulatory pathways are linked with male sterility in the TGMS line during cold stress.
Figures



References
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 - PubMed
-
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

