Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria
- PMID: 22509013
- PMCID: PMC3356625
- DOI: 10.1073/pnas.1119226109
Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria
Abstract
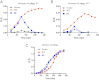
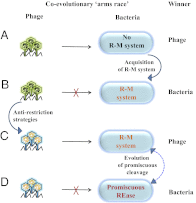
Most bacterial genomes harbor restriction-modification systems, encoding a REase and its cognate MTase. On attack by a foreign DNA, the REase recognizes it as nonself and subjects it to restriction. Should REases be highly specific for targeting the invading foreign DNA? It is often considered to be the case. However, when bacteria harboring a promiscuous or high-fidelity variant of the REase were challenged with bacteriophages, fitness was maximal under conditions of catalytic promiscuity. We also delineate possible mechanisms by which the REase recognizes the chromosome as self at the noncanonical sites, thereby preventing lethal dsDNA breaks. This study provides a fundamental understanding of how bacteria exploit an existing defense system to gain fitness advantage during a host-parasite coevolutionary "arms race."
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Transcriptome analyses of cells carrying the Type II Csp231I restriction-modification system reveal cross-talk between two unrelated transcription factors: C protein and the Rac prophage repressor.Nucleic Acids Res. 2019 Oct 10;47(18):9542-9556. doi: 10.1093/nar/gkz665. Nucleic Acids Res. 2019. PMID: 31372643 Free PMC article.
-
Ca(2+)-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease.J Biol Chem. 2004 Nov 26;279(48):49736-40. doi: 10.1074/jbc.M409483200. Epub 2004 Sep 16. J Biol Chem. 2004. PMID: 15375161
-
A mobile restriction-modification system provides phage defence and resolves an epigenetic conflict with an antagonistic endonuclease.Nucleic Acids Res. 2022 Apr 8;50(6):3348-3361. doi: 10.1093/nar/gkac147. Nucleic Acids Res. 2022. PMID: 35286398 Free PMC article.
-
Biology of DNA restriction.Microbiol Rev. 1993 Jun;57(2):434-50. doi: 10.1128/mr.57.2.434-450.1993. Microbiol Rev. 1993. PMID: 8336674 Free PMC article. Review.
-
Role of Restriction-Modification Systems in Prokaryotic Evolution and Ecology.Biochemistry (Mosc). 2015 Oct;80(10):1373-86. doi: 10.1134/S0006297915100193. Biochemistry (Mosc). 2015. PMID: 26567582 Review.
Cited by
-
Active site plasticity enables metal-dependent tuning of Cas5d nuclease activity in CRISPR-Cas type I-C system.Nucleic Acids Res. 2014 Apr;42(6):3846-56. doi: 10.1093/nar/gkt1335. Epub 2013 Dec 25. Nucleic Acids Res. 2014. PMID: 24371266 Free PMC article.
-
Naturally-occurring, dually-functional fusions between restriction endonucleases and regulatory proteins.BMC Evol Biol. 2013 Oct 2;13:218. doi: 10.1186/1471-2148-13-218. BMC Evol Biol. 2013. PMID: 24083337 Free PMC article.
-
Low-level expression of the Type II restriction-modification system confers potent bacteriophage resistance in Escherichia coli.DNA Res. 2020 Feb 1;27(1):dsaa003. doi: 10.1093/dnares/dsaa003. DNA Res. 2020. PMID: 32167561 Free PMC article.
-
Anti-Restriction Protein, KlcAHS, Promotes Dissemination of Carbapenem Resistance.Front Cell Infect Microbiol. 2017 May 2;7:150. doi: 10.3389/fcimb.2017.00150. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28512626 Free PMC article.
-
Evolutionary transitions to new DNA methyltransferases through target site expansion and shrinkage.Nucleic Acids Res. 2012 Dec;40(22):11627-37. doi: 10.1093/nar/gks944. Epub 2012 Oct 15. Nucleic Acids Res. 2012. PMID: 23074188 Free PMC article.
References
-
- Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. - PubMed
-
- Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. - PubMed
-
- O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. - PubMed
-
- Aharoni A, et al. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. - PubMed
-
- Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440:1078–1082. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

