MHC class I antigen processing distinguishes endogenous antigens based on their translation from cellular vs. viral mRNA
- PMID: 22509014
- PMCID: PMC3344963
- DOI: 10.1073/pnas.1112387109
MHC class I antigen processing distinguishes endogenous antigens based on their translation from cellular vs. viral mRNA
Abstract
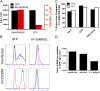
To better understand the generation of MHC class I-associated peptides, we used a model antigenic protein whose proteasome-mediated degradation is rapidly and reversibly controlled by Shield-1, a cell-permeant drug. When expressed from a stably transfected gene, the efficiency of antigen presentation is ~2%, that is, one cell-surface MHC class I-peptide complex is generated for every 50 folded source proteins degraded upon Shield-1 withdrawal. By contrast, when the same protein is expressed by vaccinia virus, its antigen presentation efficiency is reduced ~10-fold to values similar to those reported for other vaccinia virus-encoded model antigens. Virus infection per se does not modify the efficiency of antigen processing. Rather, the efficiency difference between cellular and virus-encoded antigens is based on whether the antigen is synthesized from transgene- vs. virus-encoded mRNA. Thus, class I antigen-processing machinery can distinguish folded proteins based on the precise details of their synthesis to modulate antigen presentation efficiency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

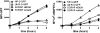
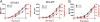
References
-
- Boon T, et al. Genes coding for T-cell-defined tum transplantation antigens: Point mutations, antigenic peptides, and subgenic expression. Cold Spring Harb Symp Quant Biol. 1989;54:587–596. - PubMed
-
- Villanueva MS, Fischer P, Feen K, Pamer EG. Efficiency of MHC class I antigen processing: A quantitative analysis. Immunity. 1994;1:479–489. - PubMed
-
- Villanueva MS, Sijts AJ, Pamer EG. Listeriolysin is processed efficiently into an MHC class I-associated epitope in Listeria monocytogenes-infected cells. J Immunol. 1995;155:5227–5233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

