Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1
- PMID: 22509017
- PMCID: PMC3344969
- DOI: 10.1073/pnas.1120174109
Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1
Abstract
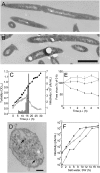
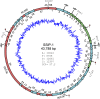
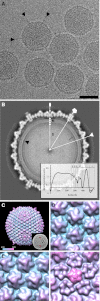
The multitude of archaea and bacteria inhabiting extreme environments has only become evident during the last decades. As viruses apply a significant evolutionary force to their hosts, there is an inherent value in learning about viruses infecting these extremophiles. In this study, we have focused on one such unique virus-host pair isolated from a hypersaline environment: an icosahedral, membrane-containing double-stranded DNA virus--Salisaeta icosahedral phage 1 (SSIP-1) and its halophilic host bacterium Salisaeta sp. SP9-1 closely related to Salisaeta longa. The architectural principles, virion composition, and the proposed functions associated with some of the ORFs of the virus are surprisingly similar to those found in viruses belonging to the PRD1-adenovirus lineage. The virion structure, determined by electron cryomicroscopy, reveals that the bulk of the outer protein capsid is composed of upright standing pseudohexameric capsomers organized on a T = 49 icosahedral lattice. Our results give a comprehensive description of a halophilic virus-host system and shed light on the relatedness of viruses based on their virion architecture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bamford DH. Do viruses form lineages across different domains of life? Res Microbiol. 2003;154:231–236. - PubMed
-
- Jalasvuori M, Bamford JK. Structural co-evolution of viruses and cells in the primordial world. Orig Life Evol Biosph. 2008;38:165–181. - PubMed
-
- Forterre P, Prangishvili D. The origin of viruses. Res Microbiol. 2009;160:466–472. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

