Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca
- PMID: 22509035
- PMCID: PMC3345007
- DOI: 10.1073/pnas.1120788109
Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca
Abstract
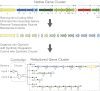
Bacterial genes associated with a single trait are often grouped in a contiguous unit of the genome known as a gene cluster. It is difficult to genetically manipulate many gene clusters because of complex, redundant, and integrated host regulation. We have developed a systematic approach to completely specify the genetics of a gene cluster by rebuilding it from the bottom up using only synthetic, well-characterized parts. This process removes all native regulation, including that which is undiscovered. First, all noncoding DNA, regulatory proteins, and nonessential genes are removed. The codons of essential genes are changed to create a DNA sequence as divergent as possible from the wild-type (WT) gene. Recoded genes are computationally scanned to eliminate internal regulation. They are organized into operons and placed under the control of synthetic parts (promoters, ribosome binding sites, and terminators) that are functionally separated by spacer parts. Finally, a controller consisting of genetic sensors and circuits regulates the conditions and dynamics of gene expression. We applied this approach to an agriculturally relevant gene cluster from Klebsiella oxytoca encoding the nitrogen fixation pathway for converting atmospheric N(2) to ammonia. The native gene cluster consists of 20 genes in seven operons and is encoded in 23.5 kb of DNA. We constructed a "refactored" gene cluster that shares little DNA sequence identity with WT and for which the function of every genetic part is defined. This work demonstrates the potential for synthetic biology tools to rewrite the genetics encoding complex biological functions to facilitate access, engineering, and transferability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Synthetic biology: We can rebuild you.Nat Rev Microbiol. 2012 May 14;10(6):378. doi: 10.1038/nrmicro2803. Nat Rev Microbiol. 2012. PMID: 22580366 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

