Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation
- PMID: 22509135
- PMCID: PMC3317911
- DOI: 10.1371/journal.pbio.1001301
Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation
Abstract
Cells employ multiple levels of regulation, including transcriptional and translational regulation, that drive core biological processes and enable cells to respond to genetic and environmental changes. Small-molecule metabolites are one category of critical cellular intermediates that can influence as well as be a target of cellular regulations. Because metabolites represent the direct output of protein-mediated cellular processes, endogenous metabolite concentrations can closely reflect cellular physiological states, especially when integrated with other molecular-profiling data. Here we develop and apply a network reconstruction approach that simultaneously integrates six different types of data: endogenous metabolite concentration, RNA expression, DNA variation, DNA-protein binding, protein-metabolite interaction, and protein-protein interaction data, to construct probabilistic causal networks that elucidate the complexity of cell regulation in a segregating yeast population. Because many of the metabolites are found to be under strong genetic control, we were able to employ a causal regulator detection algorithm to identify causal regulators of the resulting network that elucidated the mechanisms by which variations in their sequence affect gene expression and metabolite concentrations. We examined all four expression quantitative trait loci (eQTL) hot spots with colocalized metabolite QTLs, two of which recapitulated known biological processes, while the other two elucidated novel putative biological mechanisms for the eQTL hot spots.
Conflict of interest statement
I have read the journal's policy and have the following conflicts. The work was partially funded by Merck.
Figures



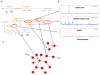


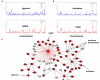

Comment in
-
Bioinformatics: Next-generation omics.Nat Rev Genet. 2012 May 2;13(6):378. doi: 10.1038/nrg3250. Nat Rev Genet. 2012. PMID: 22549153 No abstract available.
References
-
- Emilsson V, Thorleifsson G, Zhang B, Leonardson A. S, Zink F, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. - PubMed
-
- Hsu Y. H, Zillikens M. C, Wilson S. G, Farber C. R, Demissie S, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 6:e1000977. doi: 10.1371/journal.pgen.1000977. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

