Antibiotic resistance is prevalent in an isolated cave microbiome
- PMID: 22509370
- PMCID: PMC3324550
- DOI: 10.1371/journal.pone.0034953
Antibiotic resistance is prevalent in an isolated cave microbiome
Abstract
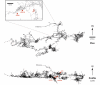
Antibiotic resistance is a global challenge that impacts all pharmaceutically used antibiotics. The origin of the genes associated with this resistance is of significant importance to our understanding of the evolution and dissemination of antibiotic resistance in pathogens. A growing body of evidence implicates environmental organisms as reservoirs of these resistance genes; however, the role of anthropogenic use of antibiotics in the emergence of these genes is controversial. We report a screen of a sample of the culturable microbiome of Lechuguilla Cave, New Mexico, in a region of the cave that has been isolated for over 4 million years. We report that, like surface microbes, these bacteria were highly resistant to antibiotics; some strains were resistant to 14 different commercially available antibiotics. Resistance was detected to a wide range of structurally different antibiotics including daptomycin, an antibiotic of last resort in the treatment of drug resistant Gram-positive pathogens. Enzyme-mediated mechanisms of resistance were also discovered for natural and semi-synthetic macrolide antibiotics via glycosylation and through a kinase-mediated phosphorylation mechanism. Sequencing of the genome of one of the resistant bacteria identified a macrolide kinase encoding gene and characterization of its product revealed it to be related to a known family of kinases circulating in modern drug resistant pathogens. The implications of this study are significant to our understanding of the prevalence of resistance, even in microbiomes isolated from human use of antibiotics. This supports a growing understanding that antibiotic resistance is natural, ancient, and hard wired in the microbial pangenome.
Conflict of interest statement
Figures




References
-
- Hughes VM, Datta N. Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature. 1983;302:725–726. - PubMed
-
- Davis CE, Anandan J. The evolution of r factor. A study of a “preantibiotic” community in Borneo. N Engl J Med. 1970;282:117–122. - PubMed
-
- Knapp CW, Dolfing J, Ehlert PA, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010;44:580–587. - PubMed
-
- Barlow M, Hall BG. Phylogenetic analysis shows that the OXA beta-lactamase genes have been on plasmids for millions of years. J Mol Evol. 2002;55:314–321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

