Structural basis of heparin binding to camel peptidoglycan recognition protein-S
- PMID: 22509483
- PMCID: PMC3325777
Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Erratum in
- Int J Biochem Mol Biol. 2013;4(4):215
Abstract
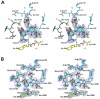
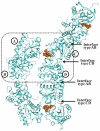
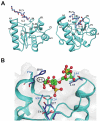
Short peptidoglycan recognition protein (PGRP-S) is a member of the innate immunity system in mammals. PGRP-S from Camelus dromedarius (CPGRP-S) is found to be highly potent against bacterial infections. It is capable of binding to a wide range of pathogen-associated molecular patterns (PAMPs) including lipopolysaccharide (LPS), lipoteichoic acid (LTA) and peptidoglycan (PGN). The heparin-like polysaccharides have also been observed in some bacteria such as the capsule of K5 Escherichia coli thus making them relevant for determining the nature of their interactions with CPGRP-S. The binding studies of CPGRP-S with heparin disaccharide in solution using surface plasmon resonance gave a value 3.3×10(-7) M for the dissociation constant (Kd). The structure of the heparin bound CPGRP-S determined at 2.8Å resolution revealed the presence of a bound heparin molecule in the binding pocket of CPGRP-S. It was found anchored tightly to the protein with the help of several ionic and hydrogen bonded interactions. Three sulphate groups of heparin S1, S2 and S3 have been found to interact with residues, Arg-31, Lys-90, Thr- 97, Asn-99 Asn-140, Gln-150 and Arg-170 of CPGRP-S. The binding site includes two subsites, S-I and S-II with cleft-like structures. Heparin disaccharide is bound in subsite S-I. Previously determined structures of the complexes of CPGRP-S with LPS, LTA and PGN also showed that their glycan moieties were also held in subsite S-I indicating that heparin disaccharide also represents an important element for the recognition by CPGRP-S.
Keywords: LPS; LTA; PAMPs; PGN; PGRP-S; crystal structure; heparin.
Figures

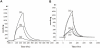



Similar articles
-
Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.PLoS One. 2013;8(1):e53756. doi: 10.1371/journal.pone.0053756. Epub 2013 Jan 9. PLoS One. 2013. PMID: 23326499 Free PMC article.
-
Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein.J Biol Chem. 2011 May 6;286(18):16208-17. doi: 10.1074/jbc.M111.228163. Epub 2011 Mar 21. J Biol Chem. 2011. PMID: 21454594 Free PMC article.
-
Structural studies on molecular interactions between camel peptidoglycan recognition protein, CPGRP-S, and peptidoglycan moieties N-acetylglucosamine and N-acetylmuramic acid.J Biol Chem. 2012 Jun 22;287(26):22153-64. doi: 10.1074/jbc.M111.321307. Epub 2012 May 9. J Biol Chem. 2012. Retraction in: J Biol Chem. 2014 Aug 22;289(34):23331. doi: 10.1074/jbc.A111.321307. PMID: 22573327 Free PMC article. Retracted.
-
Roles of peptidoglycan recognition protein (PGRP) in immunity and implications for novel anti-infective measures.Crit Rev Eukaryot Gene Expr. 2012;22(3):259-68. doi: 10.1615/critreveukargeneexpr.v22.i3.90. Crit Rev Eukaryot Gene Expr. 2012. PMID: 23140167 Review.
-
Pulmonary innate immune proteins and receptors that interact with gram-positive bacterial ligands.Immunobiology. 2002 Sep;205(4-5):575-94. doi: 10.1078/0171-2985-00156. Immunobiology. 2002. PMID: 12396017 Review.
Cited by
-
Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.PLoS One. 2013;8(1):e53756. doi: 10.1371/journal.pone.0053756. Epub 2013 Jan 9. PLoS One. 2013. PMID: 23326499 Free PMC article.
-
Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 Å resolution.Int J Biochem Mol Biol. 2022 Aug 20;13(4):28-39. eCollection 2022. Int J Biochem Mol Biol. 2022. PMID: 36188729 Free PMC article.
-
Structural similarities in the CPC clip motif explain peptide-binding promiscuity between glycosaminoglycans and lipopolysaccharides.J R Soc Interface. 2017 Nov;14(136):20170423. doi: 10.1098/rsif.2017.0423. J R Soc Interface. 2017. PMID: 29187635 Free PMC article.
References
-
- Medzhitov R, Janeway C Jr. Innate immunity. N Engl J Med. 2000;343:338–344. - PubMed
-
- Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. - PubMed
-
- Kappeler SR, Heuberger C, Farah Z, Puhan Z. Expression of the peptidoglycan recognition protein, PGRP, in the lactating mammary gland. J Dairy Sci. 2004;87:2660–2668. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
