FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c
- PMID: 22511764
- PMCID: PMC3370196
- DOI: 10.1074/jbc.M112.347211
FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c
Abstract
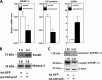
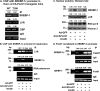
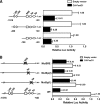
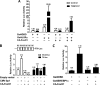
Induction of lipogenesis in response to insulin is critically dependent on the transcription factor, sterol regulatory element-binding protein-1c (SREBP-1c). FoxO1, a forkhead box class-O transcription factor, is an important mediator of insulin action, but its role in the regulation of lipid metabolism has not been clearly defined. We examined the effects of FoxO1 on srebp1 gene expression in vivo and in vitro. In vivo studies showed that constitutively active (CA) FoxO1 (CA-FoxO1) reduced basal expression of SREBP-1c mRNA in liver by ∼60% and blunted induction of SREBP-1c in response to feeding. In liver-specific FoxO knock-out mice, SREBP-1c expression was increased ∼2-fold. Similarly, in primary hepatocytes, CA-FoxO1 suppressed SREBP1-c expression and inhibited basal and insulin-induced SREBP-1c promoter activity. SREBP-1c gene expression is induced by the liver X receptor (LXR), but CA-FoxO1 did not block the activation of SREBP-1c by the LXR agonist TO9. Insulin stimulates SREBP-1c transcription through Sp1 and via "feed forward" regulation by newly synthesized SREBP-1c. CA-FoxO1 inhibited SREBP-1c by reducing the transactivational capacity of both Sp1 and SREBP-1c. In addition, chromatin immunoprecipitation assays indicate that FoxO1 can associate with the proximal promoter region of the srebp1 gene and disrupt the assembly of key components of the transcriptional complex of the SREBP-1c promoter. We conclude that FoxO1 inhibits SREBP-1c transcription via combined actions on multiple transcription factors and that this effect is exerted at least in part through reduced transcriptional activity of Sp1 and SREBP-1c and disrupted assembly of the transcriptional initiation complex on the SREBP-1c promoter.
Figures



References
-
- Barthel A., Schmoll D., Unterman T. G. (2005) FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 16, 183–189 - PubMed
-
- Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 - PubMed
-
- Raghow R., Yellaturu C., Deng X., Park E. A., Elam M. B. (2008) SREBPs. The cross-roads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 19, 65–73 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

