Effects of BRCA2 cis-regulation in normal breast and cancer risk amongst BRCA2 mutation carriers
- PMID: 22513257
- PMCID: PMC3446398
- DOI: 10.1186/bcr3169
Effects of BRCA2 cis-regulation in normal breast and cancer risk amongst BRCA2 mutation carriers
Abstract
Introduction: Cis-acting regulatory single nucleotide polymorphisms (SNPs) at specific loci may modulate penetrance of germline mutations at the same loci by introducing different levels of expression of the wild-type allele. We have previously reported that BRCA2 shows differential allelic expression and we hypothesize that the known variable penetrance of BRCA2 mutations might be associated with this mechanism.
Methods: We combined haplotype analysis and differential allelic expression of BRCA2 in breast tissue to identify expression haplotypes and candidate cis-regulatory variants. These candidate variants underwent selection based on in silico predictions for regulatory potential and disruption of transcription factor binding, and were functionally analyzed in vitro and in vivo in normal and breast cancer cell lines. SNPs tagging the expression haplotypes were correlated with the total expression of several genes in breast tissue measured by Taqman and microarray technologies. The effect of the expression haplotypes on breast cancer risk in BRCA2 mutation carriers was investigated in 2,754 carriers.
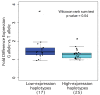
Results: We identified common haplotypes associated with differences in the levels of BRCA2 expression in human breast cells. We characterized three cis-regulatory SNPs located at the promoter and two intronic regulatory elements which affect the binding of the transcription factors C/EBPα, HMGA1, D-binding protein (DBP) and ZF5. We showed that the expression haplotypes also correlated with changes in the expression of other genes in normal breast. Furthermore, there was suggestive evidence that the minor allele of SNP rs4942440, which is associated with higher BRCA2 expression, is also associated with a reduced risk of breast cancer (per-allele hazard ratio (HR) = 0.85, 95% confidence interval (CI) = 0.72 to 1.00, P-trend = 0.048).
Conclusions: Our work provides further insights into the role of cis-regulatory variation in the penetrance of disease-causing mutations. We identified small-effect genetic variants associated with allelic expression differences in BRCA2 which could possibly affect the risk in mutation carriers through altering expression levels of the wild-type allele.
Figures




References
-
- Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, Zeng Q, Zhang H-T, Pennison MJ, Sadim M, Pasche B, Tanner SM, de la Chapelle A. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. - DOI - PMC - PubMed
-
- Tan AC, Fan J-B, Karikari C, Bibikova M, Garcia EW, Zhou L, Barker D, Serre D, Feldmann G, Hruban RH, Klein AP, Goggins M, Couch FJ, Hudson TJ, Winslow RL, Maitra A, Chakravarti A. Allele-specific expression in the germline of patients with familial pancreatic cancer: an unbiased approach to cancer gene discovery. Cancer Biol Ther. 2008;7:135–144. doi: 10.4161/cbt.7.1.5199. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

