Novel APP/Aβ mutation K16N produces highly toxic heteromeric Aβ oligomers
- PMID: 22514144
- PMCID: PMC3407951
- DOI: 10.1002/emmm.201200239
Novel APP/Aβ mutation K16N produces highly toxic heteromeric Aβ oligomers
Abstract
Here, we describe a novel missense mutation in the amyloid precursor protein (APP) causing a lysine-to-asparagine substitution at position 687 (APP770; herein, referred to as K16N according to amyloid-β (Aβ) numbering) resulting in an early onset dementia with an autosomal dominant inheritance pattern. The K16N mutation is located exactly at the α-secretase cleavage site and influences both APP and Aβ. First, due to the K16N mutation APP secretion is affected and a higher amount of Aβ peptides is being produced. Second, Aβ peptides carrying the K16N mutation are unique in that the peptide itself is not harmful to neuronal cells. Severe toxicity, however, is evident upon equimolar mixture of wt and mutant peptides, mimicking the heterozygous state of the subject. Furthermore, Aβ42 K16N inhibits fibril formation of Aβ42 wild-type. Even more, Aβ42 K16N peptides are protected against clearance activity by the major Aβ-degrading enzyme neprilysin. Thus the mutation characterized here harbours a combination of risk factors that synergistically may contribute to the development of early onset Alzheimer disease.
Copyright © 2012 EMBO Molecular Medicine.
Figures

Schematic of APP processing. FL-APP is initially cleaved by either the α- or β-secretase. The remaining membrane-bound C-terminal fragments α-CTF and β-CTF, respectively, are further cleaved by the γ-secretase to generate the APP intracellular domain (AICD) and p3 or Aβ. Aβ region is shown in grey. Part of the APP sequence highlighting the Aβ42 region (underlined). The mutation at the α-secretase cleavage site changes the lysine in position 687 of APP (according to APP770 numbering) or position 16 of Aβ to asparagine. Known secretase-cleavage sites are marked by arrows. β′ is an alternative β-secretase cleavage site.
Three-generation pedigree of the family. The index patient is marked with an arrow. Four of six siblings of generation II suffered from dementia. Siblings of the index patient have no dementia and have not been tested for the mutation.
Aβ from the patient's CSF was precipitated with the monoclonal antibody W0-2 and Aβ masses were determined by MALDI-MS. In CSF of the patient with the K16N mutation a mass shift of 14 Da was detected between wt and mutant Aβ, indicated as Aβ37/38/39/40 (wt) and Aβ37/38/39/40* (K16N). Only in the CSF from the patient suffering from alcoholic dementia Aβ42 was detected. See Table S2 of Supporting Information for the experimental and theoretical masses.
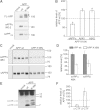
Representative Western blots of FL-APP (mature forms are marked with *) and soluble APP (sAPPtotal). FL-APP and sAPPtotal were detected with the myc- and sAPPα by W0-2 antibody. The empty vector (mock) was used as a control.
Quantification of sAPPα, Aβ40 and Aβ42 levels by ELISA. Shown are the respective levels of proteolytic fragments normalized to 100% of APP wt (n = 4 ± SEM). Asterisks indicate significant differences between mutant and wt APP (*p < 0.05 one-way ANOVA followed by posthoc Dunnett).
Representative Western blots of sAPPα detected with the 4B4 antibody (Kuhn et al, 2010) and sAPPβ detected with the anti-sAPPβ antibody (IBL).
Densitometric quantification of sAPPα and sAPPβ levels from Western blot analysis (five-independent Western Blots with 4–6 samples each).
Representative Western blot of APP-CTFs. Lysates were subjected to immunprecipitation and analysed by an APP-C-terminal antibody.
Densitometric quantification of the levels of α-CTF and β-CTF from Western blot analysis. Shown are the respective levels of CTFs normalized to 100% of APP wt (n = 3).

A-C. Mass spectra of trypsin and ADAM10 proteolysis of Aβ11–28 peptides: wt (A), K16N (B), E22G (arctic) (C). Peptides were incubated for the indicated times with ADAM10 or trypsin and mixed with α-cyano-4-hydroxycinnamic acid matrix and analysed by MALDI-MS. Peptide identities are indicated at the top. See Figure S6 and Table S4 of Supporting Information for details and the experimental and predicted masses. Note, Aβ11–28 K16N is not cleaved by trypsin and cleavage by ADAM10 is significantly slowed down.
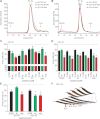
A,B. SEC of freshly dissolved Aβ40 or Aβ42 wt (green), K16N (black) and the equimolar mixture of both peptides (mix, red). The chromatograms show all similar distributions of low molecular weight oligomers. Note that in the Aβ42 mix, the oligomer distribution shifted from low-n oligomers (4–6x) to high-n oligomers (16–20x).
C,D. SH-SY5Y cells were incubated for 12 h with either 2 µM freshly dissolved peptides (load) or oligomers (2–20x) obtained by SEC.
E. Primary hippocampal neurons were incubated for 48 h with 2 µM freshly dissolved peptides. In C–E toxicity was determined by percentage of living cells compared to untreated control cells (n = 4–8 +SEM). One-way ANOVA, Bonferroni's multiple comparison test (*p < 0.001 and **p < 0.0001).
F. Tertiary structure model for Aβ1–42 mix (wt + K16N) tetramer based on the solution NMR structure Luhrs et al (2005) (see methods). In this model, K16 and N16 form a 2.0 Å short, and thus presumably strong, inter-strand hydrogen bond (black dots). The side chains of K16 and N16 are depicted as sticks, with the nitrogen and oxygen atoms in red and blue, respectively.
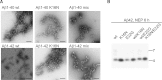
Aggregation properties of Aβ wt and K16N peptides. Electron micrographs of freshly dissolved Aβ40 or Aβ42 peptides aged for 24 h and negatively stained. Mix indicates equimolar mixtures of wt and K16N peptides. Only Aβ42 wt peptides form long and rigid fibrils, whereas, co-aggregation of wt and K16N peptides leads to the formation of aggregates comparable with K16N alone. Bar 100 nm.
Freshly dissolved Aβ42 peptides (wt, K16N, E22G and the respective 1:1 mixes wt/K16N, wt/E22G and K16N/E22G) were incubated for 6 h with human NEP and analysed by Western blot analysis. Aβ was detected by the W0-2 antibody. Aβ42 K16N peptides are more stable against NEP proteolysis.

Spectra with mass range from 700 to 1070 Da.
Spectra with mass range from 1920 to 2070 Da.
References
-
- Carson JA, Turner AJ. Beta-amyloid catabolism: Roles for neprilysin (NEP) and other metallopeptidases. J Neurochem. 2002;81:1–8. - PubMed
-
- Chen YR, Glabe CG. Distinct early folding and aggregation properties of Alzheimer amyloid-beta peptides Abeta40 and Abeta42: stable trimer or tetramer formation by Abeta42. J Biol Chem. 2006;281:24414–24422. - PubMed
-
- De Jonghe C, Zehr C, Yager D, Prada CM, Younkin S, Hendriks L, Van Broeckhoven C, Eckman CB. Flemish and Dutch mutations in amyloid beta precursor protein have different effects on amyloid beta secretion. Neurobiol Dis. 1998;5:281–286. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

