Cooperation of nuclear fibroblast growth factor receptor 1 and Nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons
- PMID: 22514272
- PMCID: PMC3370168
- DOI: 10.1074/jbc.M112.347831
Cooperation of nuclear fibroblast growth factor receptor 1 and Nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons
Abstract
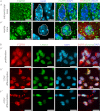
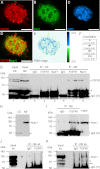
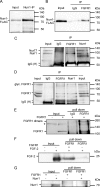
Experiments in mice deficient for Nurr1 or expressing the dominant-negative FGF receptor (FGFR) identified orphan nuclear receptor Nurr1 and FGFR1 as essential factors in development of mesencephalic dopaminergic (mDA) neurons. FGFR1 affects brain cell development by two distinct mechanisms. Activation of cell surface FGFR1 by secreted FGFs stimulates proliferation of neural progenitor cells, whereas direct integrative nuclear FGFR1 signaling (INFS) is associated with an exit from the cell cycle and neuronal differentiation. Both Nurr1 and INFS activate expression of neuronal genes, such as tyrosine hydroxylase (TH), which is the rate-limiting enzyme in dopamine synthesis. Here, we show that nuclear FGFR1 and Nurr1 are expressed in the nuclei of developing TH-positive cells in the embryonic ventral midbrain. Both nuclear receptors were effectively co-immunoprecipitated from the ventral midbrain of FGF-2-deficient embryonic mice, which previously showed an increase of mDA neurons and enhanced nuclear FGFR1 accumulation. Immunoprecipitation and co-localization experiments showed the presence of Nurr1 and FGFR1 in common nuclear protein complexes. Fluorescence recovery after photobleaching and chromatin immunoprecipitation experiments demonstrated the Nurr1-mediated shift of nuclear FGFR1-EGFP mobility toward a transcriptionally active population and that both Nurr1 and FGFR1 bind to a common region in the TH gene promoter. Furthermore, nuclear FGFR1 or its 23-kDa FGF-2 ligand (FGF-2(23)) enhances Nurr1-dependent activation of the TH gene promoter. Transcriptional cooperation of FGFR1 with Nurr1 was confirmed on isolated Nurr1-binding elements. The proposed INFS/Nurr1 nuclear partnership provides a novel mechanism for TH gene regulation in mDA neurons and a potential therapeutic target in neurodevelopmental and neurodegenerative disorders.
Figures



References
-
- Klejbor I., Myers J. M., Hausknecht K., Corso T. D., Gambino A. S., Morys J., Maher P. A., Hard R., Richards J., Stachowiak E. K., Stachowiak M. K. (2006) Fibroblast growth factor receptor signaling affects development and function of dopamine neurons. Inhibition results in a schizophrenia-like syndrome in transgenic mice. J. Neurochem. 97, 1243–1258 - PubMed
-
- Smidt M. P., Burbach J. P. (2009) Terminal differentiation ofmesodiencephalic dopaminergic neurons. The role of Nurr1 and Pitx3. Adv. Exp. Med. Biol. 651, 47–57 - PubMed
-
- Law S. W., Conneely O. M., DeMayo F. J., O'Malley B. W. (1992) Identification of a new brain-specific transcription factor, NURR1. Mol. Endocrinol. 6, 2129–2135 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

