Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II
- PMID: 22514352
- PMCID: PMC3416352
- DOI: 10.1128/JVI.00541-12
Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II
Abstract
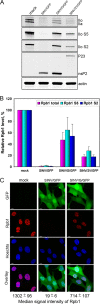
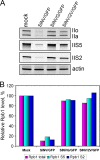
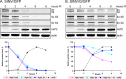
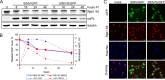
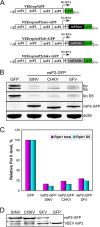
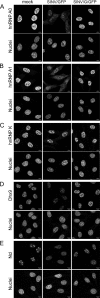
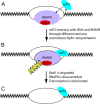
The Old World alphaviruses are emerging human pathogens with an ability to cause widespread epidemics. The latest epidemic of Chikungunya virus, from 2005 to 2007, affected over 40 countries in Africa, Asia, and Europe. The Old World alphaviruses are highly cytopathic and known to evade the cellular antiviral response by inducing global inhibition of transcription in vertebrate cells. This function was shown to be mediated by their nonstructural nsP2 protein; however, the detailed mechanism of this phenomenon has remained unknown. Here, we report that nsP2 proteins of Sindbis, Semliki Forest, and Chikungunya viruses inhibit cellular transcription by inducing rapid degradation of Rpb1, a catalytic subunit of the RNAPII complex. This degradation of Rpb1 is independent of the nsP2-associated protease activity, but, instead, it proceeds through nsP2-mediated Rpb1 ubiquitination. This function of nsP2 depends on the integrity of the helicase and S-adenosylmethionine (SAM)-dependent methyltransferase-like domains, and point mutations in either of these domains abolish Rpb1 degradation. We go on to show that complete degradation of Rpb1 in alphavirus-infected cells occurs within 6 h postinfection, before other previously described virus-induced changes in cell physiology, such as apoptosis, autophagy, and inhibition of STAT1 phosphorylation, are detected. Since Rpb1 is a subunit that catalyzes the polymerase reaction during RNA transcription, degradation of Rpb1 plays an indispensable role in blocking the activation of cellular genes and downregulating cellular antiviral response. This indicates that the nsP2-induced degradation of Rpb1 is a critical mechanism utilized by the Old World alphaviruses to subvert the cellular antiviral response.
Figures



References
-
- Anindya R, Aygun O, Svejstrup JQ. 2007. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell 28:386–397 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

