Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects
- PMID: 22515365
- PMCID: PMC3351742
- DOI: 10.1186/1742-4690-9-30
Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects
Abstract
Background: Disassembly of the viral capsid following penetration into the cytoplasm, or uncoating, is a poorly understood stage of retrovirus infection. Based on previous studies of HIV-1 CA mutants exhibiting altered capsid stability, we concluded that formation of a capsid of optimal intrinsic stability is crucial for HIV-1 infection.
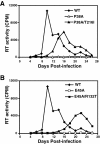
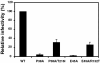
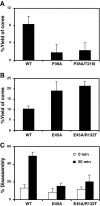
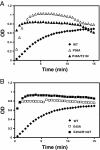
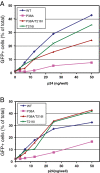
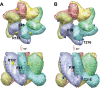
Results: To further examine the connection between HIV-1 capsid stability and infectivity, we isolated second-site suppressors of HIV-1 mutants exhibiting unstable (P38A) or hyperstable (E45A) capsids. We identified the respective suppressor mutations, T216I and R132T, which restored virus replication in a human T cell line and markedly enhanced the fitness of the original mutants as revealed in single-cycle infection assays. Analysis of the corresponding purified N-terminal domain CA proteins by NMR spectroscopy demonstrated that the E45A and R132T mutations induced structural changes that are localized to the regions of the mutations, while the P38A mutation resulted in changes extending to neighboring regions in space. Unexpectedly, neither suppressor mutation corrected the intrinsic viral capsid stability defect associated with the respective original mutation. Nonetheless, the R132T mutation rescued the selective infectivity impairment exhibited by the E45A mutant in aphidicolin-arrested cells, and the double mutant regained sensitivity to the small molecule inhibitor PF74. The T216I mutation rescued the impaired ability of the P38A mutant virus to abrogate restriction by TRIMCyp and TRIM5α.
Conclusions: The second-site suppressor mutations in CA that we have identified rescue virus infection without correcting the intrinsic capsid stability defects associated with the P38A and E45A mutations. The suppressors also restored wild type virus function in several cell-based assays. We propose that while proper HIV-1 uncoating in target cells is dependent on the intrinsic stability of the viral capsid, the effects of stability-altering mutations can be mitigated by additional mutations that affect interactions with host factors in target cells or the consequences of these interactions. The ability of mutations at other CA surfaces to compensate for effects at the NTD-NTD interface further indicates that uncoating in target cells is controlled by multiple intersubunit interfaces in the viral capsid.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

