Molecular characterization of rotavirus strains detected during a clinical trial of a human rotavirus vaccine in Blantyre, Malawi
- PMID: 22520123
- PMCID: PMC3982048
- DOI: 10.1016/j.vaccine.2011.09.119
Molecular characterization of rotavirus strains detected during a clinical trial of a human rotavirus vaccine in Blantyre, Malawi
Abstract
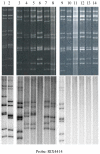
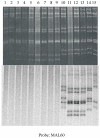
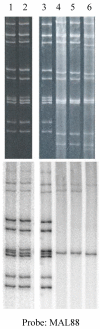
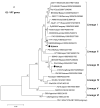
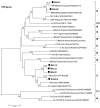
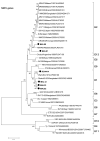
The human, G1P[8] rotavirus vaccine (Rotarix™) significantly reduced severe rotavirus gastroenteritis episodes in a clinical trial in South Africa and Malawi, but vaccine efficacy was lower in Malawi (49.5%) than reported in South Africa (76.9%) and elsewhere. The aim of this study was to examine the molecular relationships of circulating wild-type rotaviruses detected during the clinical trial in Malawi to RIX4414 (the strain contained in Rotarix™) and to common human rotavirus strains. Of 88 rotavirus-positive, diarrhoeal stool specimens, 43 rotaviruses exhibited identifiable RNA migration patterns when examined by polyacrylamide gel electrophoresis. The genes encoding VP7, VP4, VP6 and NSP4 of 5 representative strains possessing genotypes G12P[6], G1P[8], G9P[8], and G8P[4] were sequenced. While their VP7 (G) and VP4 (P) genotype designations were confirmed, the VP6 (I) and NSP4 (E) genotypes were either I1E1 or I2E2, indicating that they were of human rotavirus origin. RNA-RNA hybridization using 21 culture-adapted strains showed that Malawian rotaviruses had a genomic RNA constellation common to either the Wa-like or the DS-1 like human rotaviruses. Overall, the Malawi strains appear similar in their genetic make-up to rotaviruses described in countries where vaccine efficacy is greater, suggesting that the lower efficacy in Malawi is unlikely to be explained by the diversity of circulating strains.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures







References
-
- Parashar UD, Burton A, Lanata C, Boschi-Pinto C, Shibuya K, Steele D, et al. Global mortality associated with rotavirus disease among children in 2004. J Infect Dis. 2009;200:S9–15. - PubMed
-
- Naghipour M, Nakagomi T, Nakagomi O. Issues with reducing the rotavirus-associated mortality by vaccination in developing countries. Vaccine. 2008;26:3236–41. - PubMed
-
- Cunliffe N, Nakagomi O. Introduction of rotavirus vaccines in developing countries: remaining challenges. Ann Trop Paediatr. 2007;27:157–67. - PubMed
-
- Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. - PubMed
-
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

