Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans
- PMID: 22520464
- PMCID: PMC3334877
- DOI: 10.1016/j.chom.2012.02.007
Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans
Abstract
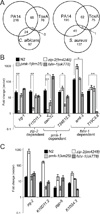
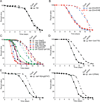
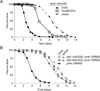
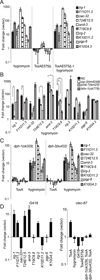
Intestinal epithelial cells are exposed to both innocuous and pathogenic microbes, which need to be distinguished to mount an effective immune response. To understand the mechanisms underlying pathogen recognition, we investigated how Pseudomonas aeruginosa triggers intestinal innate immunity in Caenorhabditis elegans, a process independent of Toll-like pattern recognition receptors. We show that the P. aeruginosa translational inhibitor Exotoxin A (ToxA), which ribosylates elongation factor 2 (EF2), upregulates a significant subset of genes normally induced by P. aeruginosa. Moreover, immune pathways involving the ATF-7 and ZIP-2 transcription factors, which protect C. elegans from P. aeruginosa, are required for preventing ToxA-mediated lethality. ToxA-responsive genes are not induced by enzymatically inactive ToxA protein but can be upregulated independently of ToxA by disruption of host protein translation. Thus, C. elegans has a surveillance mechanism to recognize ToxA through its effect on protein translation rather than by direct recognition of either ToxA or ribosylated EF2.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Comment in
-
UnZIPping mechanisms of effector-triggered immunity in animals.Cell Host Microbe. 2012 Apr 19;11(4):320-2. doi: 10.1016/j.chom.2012.04.002. Cell Host Microbe. 2012. PMID: 22520459 Free PMC article.
References
-
- Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere Guillaume M, Ip WKE, Fracchia S, Hennessy E, et al. Pathogen-Derived Effectors Trigger Protective Immunity via Activation of the Rac2 Enzyme and the IMD or Rip Kinase Signaling Pathway. Immunity. 2011;35:536–549. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

