C. elegans detects pathogen-induced translational inhibition to activate immune signaling
- PMID: 22520465
- PMCID: PMC3334869
- DOI: 10.1016/j.chom.2012.02.008
C. elegans detects pathogen-induced translational inhibition to activate immune signaling
Abstract
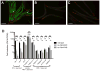
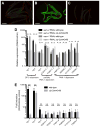
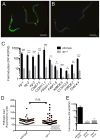
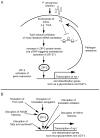
Pathogens commonly disrupt host cell processes or cause damage, but the surveillance mechanisms used by animals to monitor these attacks are poorly understood. Upon infection with pathogenic Pseudomonas aeruginosa, the nematode C. elegans upregulates infection response gene irg-1 using the zip-2 bZIP transcription factor. Here we show that P. aeruginosa infection inhibits mRNA translation in the intestine via the endocytosed translation inhibitor Exotoxin A, which leads to an increase in ZIP-2 protein levels. In the absence of infection we find that the zip-2/irg-1 pathway is upregulated following disruption of several core host processes, including inhibition of mRNA translation. ZIP-2 induction is conferred by a conserved upstream open reading frame in zip-2 that could derepress ZIP-2 translation upon infection. Thus, translational inhibition, a common pathogenic strategy, can trigger activation of an immune surveillance pathway to provide host defense.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Comment in
-
UnZIPping mechanisms of effector-triggered immunity in animals.Cell Host Microbe. 2012 Apr 19;11(4):320-2. doi: 10.1016/j.chom.2012.04.002. Cell Host Microbe. 2012. PMID: 22520459 Free PMC article.
References
-
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature immunology. 2005;6:973–979. - PubMed
-
- Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, Peters G, Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 2011 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

