Co-transcriptional degradation of aberrant pre-mRNA by Xrn2
- PMID: 22522706
- PMCID: PMC3365414
- DOI: 10.1038/emboj.2012.101
Co-transcriptional degradation of aberrant pre-mRNA by Xrn2
Abstract
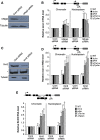
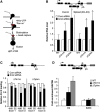
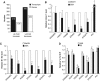
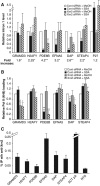
Eukaryotic protein-coding genes are transcribed as pre-mRNAs that are matured by capping, splicing and cleavage and polyadenylation. Although human pre-mRNAs can be long and complex, containing multiple introns and many alternative processing sites, they are usually processed co-transcriptionally. Mistakes during nuclear mRNA maturation could lead to potentially harmful transcripts that are important to eliminate. However, the processes of human pre-mRNA degradation are not well characterised in the human nucleus. We have studied how aberrantly processed pre-mRNAs are degraded and find a role for the 5'→3' exonuclease, Xrn2. Xrn2 associates with and co-transcriptionally degrades nascent β-globin transcripts, mutated to inhibit splicing or 3' end processing. Importantly, we provide evidence that many endogenous pre-mRNAs are also co-transcriptionally degraded by Xrn2 when their processing is inhibited by Spliceostatin A. Our data therefore establish a previously unknown function for Xrn2 and an important further aspect of pre-mRNA metabolism that occurs co-transcriptionally.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT (2002) The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841 - PubMed
-
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

