Complete DNA sequence analysis of enterohemorrhagic Escherichia coli plasmid pO157_2 in β-glucuronidase-positive E. coli O157:H7 reveals a novel evolutionary path
- PMID: 22522897
- PMCID: PMC3434749
- DOI: 10.1128/JB.00197-12
Complete DNA sequence analysis of enterohemorrhagic Escherichia coli plasmid pO157_2 in β-glucuronidase-positive E. coli O157:H7 reveals a novel evolutionary path
Abstract
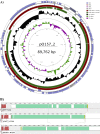
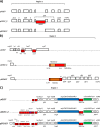
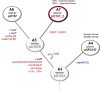
Strains of enterohemorragic Escherichia coli (EHEC) O157:H7 that are non-sorbitol fermenting (NSF) and β-glucuronidase negative (GUD(-)) carry a large virulence plasmid, pO157 (>90,000 bp), whereas closely related sorbitol-fermenting (SF) E. coli O157:H(-) strains carry plasmid pSFO157 (>120,000 bp). GUD(+) NSF O157:H7 strains are presumed to be precursors of GUD(-) NSF O157:H7 strains that also carry pO157. In this study, we report the complete sequence of a novel virulence plasmid, pO157-2 (89,762 bp), isolated from GUD(+) NSF O157:H7 strain G5101. PCR analysis confirmed the presence of pO157-2 in six other strains of GUD(+) NSF O157:H7. pO157-2 carries genes associated with virulence (e.g., hemolysin genes) and conjugation (tra and trb genes) but lacks katP and espP present in pO157. Comparative analysis of the three EHEC plasmids shows that pO157-2 is highly related to pO157 and pSFO157 but not ancestral to pO157. These results indicated that GUD(+) NSF O157:H7 strains might not be direct precursors to GUD(-) NSF O157:H7 as previously proposed but rather have evolved independently from a common ancestor.
Figures

Similar articles
-
Sorbitol-Fermenting Enterohemorrhagic Escherichia coli O157:H- Isolates from Czech Patients with Novel Plasmid Composition Not Previously Seen in German Isolates.Appl Environ Microbiol. 2017 Nov 16;83(23):e01454-17. doi: 10.1128/AEM.01454-17. Print 2017 Dec 1. Appl Environ Microbiol. 2017. PMID: 28970221 Free PMC article.
-
Sorbitol-fermenting, β-glucuronidase-positive, Shiga toxin-negative Escherichia coli O157:H7 in free-ranging red deer in South-Central Spain.Foodborne Pathog Dis. 2011 Dec;8(12):1313-5. doi: 10.1089/fpd.2011.0923. Epub 2011 Aug 5. Foodborne Pathog Dis. 2011. PMID: 21819212
-
Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H- strain 3072/96.Int J Med Microbiol. 2006 Nov;296(7):467-74. doi: 10.1016/j.ijmm.2006.05.005. Epub 2006 Aug 2. Int J Med Microbiol. 2006. PMID: 16887390
-
A brief overview of Escherichia coli O157:H7 and its plasmid O157.J Microbiol Biotechnol. 2010 Jan;20(1):5-14. J Microbiol Biotechnol. 2010. PMID: 20134227 Free PMC article. Review.
-
Pathogenicity, host responses and implications for management of enterohemorrhagic Escherichia coli O157:H7 infection.Can J Gastroenterol. 2013;27(5):281-5. doi: 10.1155/2013/138673. Can J Gastroenterol. 2013. PMID: 23712303 Free PMC article. Review.
Cited by
-
Utility of Whole-Genome Sequencing of Escherichia coli O157 for Outbreak Detection and Epidemiological Surveillance.J Clin Microbiol. 2015 Nov;53(11):3565-73. doi: 10.1128/JCM.01066-15. Epub 2015 Sep 9. J Clin Microbiol. 2015. PMID: 26354815 Free PMC article.
-
Whole-Genome Sequences of 12 Clinical Strains of Listeria monocytogenes.Genome Announc. 2015 Feb 26;3(1):e01203-14. doi: 10.1128/genomeA.01203-14. Genome Announc. 2015. PMID: 25720683 Free PMC article.
-
Genomic diversity and virulence profiles of historical Escherichia coli O157 strains isolated from clinical and environmental sources.Appl Environ Microbiol. 2015 Jan;81(2):569-77. doi: 10.1128/AEM.02616-14. Epub 2014 Nov 7. Appl Environ Microbiol. 2015. PMID: 25381234 Free PMC article.
-
Sorbitol-Fermenting Enterohemorrhagic Escherichia coli O157:H- Isolates from Czech Patients with Novel Plasmid Composition Not Previously Seen in German Isolates.Appl Environ Microbiol. 2017 Nov 16;83(23):e01454-17. doi: 10.1128/AEM.01454-17. Print 2017 Dec 1. Appl Environ Microbiol. 2017. PMID: 28970221 Free PMC article.
-
Prevalence, biogenesis, and functionality of the serine protease autotransporter EspP.Toxins (Basel). 2012 Dec 28;5(1):25-48. doi: 10.3390/toxins5010025. Toxins (Basel). 2012. PMID: 23274272 Free PMC article. Review.
References
-
- Brunder W, Karch H, Schmidt H. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− strain 3072/96. Int. J. Med. Microbiol. 296:467–474 - PubMed
-
- Brunder W, Schmidt H, Frosch M, Karch H. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005–1014 - PubMed
-
- Brunder W, Schmidt H, Karch H. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305–3315 - PubMed
-
- Brunder W, Schmidt H, Karch H. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767–778 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

