Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression
- PMID: 22523078
- PMCID: PMC3370234
- DOI: 10.1074/jbc.M112.354761
Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression
Abstract
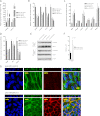
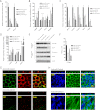
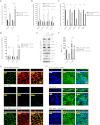
The retinal pigment epithelium (RPE) plays a fundamental role in maintaining visual function and dedifferentiation of RPE contributes to the pathophysiology of several ocular diseases. To identify microRNAs (miRNAs) that may be involved in RPE differentiation, we compared the miRNA expression profiles of differentiated primary human fetal RPE (hfRPE) cells to dedifferentiated hfRPE cells. We found that miR-204/211, the two most highly expressed miRNAs in the RPE, were significantly down-regulated in dedifferentiated hfRPE cells. Importantly, transfection of pre-miR-204/211 into hfRPE cells promoted differentiation whereas adding miR-204/211 inhibitors led to their dedifferentiation. Microphthalmia-associated transcription factor (MITF) is a key regulator of RPE differentiation that was also down-regulated in dedifferentiated hfRPE cells. MITF knockdown decreased miR-204/211 expression and caused hfRPE dedifferentiation. Significantly, co-transfection of MITF siRNA with pre-miR-204/211 rescued RPE phenotype. Collectively, our data show that miR-204/211 promote RPE differentiation, suggesting that miR-204/211-based therapeutics may be effective treatments for diseases that involve RPE dedifferentiation such as proliferative vitreoretinopathy.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

