Bacterial amyloids
- PMID: 22528099
- PMCID: PMC5324733
- DOI: 10.1007/978-1-61779-551-0_21
Bacterial amyloids
Abstract
Many bacteria can assemble functional amyloid fibers on their cell surface. The majority of bacterial amyloids contribute to biofilm or other community behaviors where cells interact with a surface or with another cell. Bacterial amyloids, like all functional amyloids, share structural and biochemical properties with disease-associated eukaryotic amyloids. The general ability of amyloids to bind amyloid-specific dyes, such as Congo red, and their resistance to denaturation have provided useful tools for scoring and quantifying bacterial amyloid formation. Here, we present basic approaches to study bacterial amyloids by focusing on the well-studied curli amyloid fibers expressed by Enterobacteriaceae. These methods exploit the specific tinctorial and biophysical properties of amyloids. The methods described here are straightforward and can be easily applied by any modern molecular biology lab for the study of other bacterial amyloids.
Figures
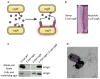
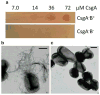


References
-
- Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebro-vascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. - PubMed
-
- Prusiner SB. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci. 1996;21:482–487. - PubMed
-
- Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wosten HA. Amyloids--a functional coat for microorganisms. Nat Rev Microbiol. 2005;3:333–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

