Yeast toxicogenomics: genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology
- PMID: 22529852
- PMCID: PMC3329712
- DOI: 10.3389/fgene.2012.00063
Yeast toxicogenomics: genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology
Abstract
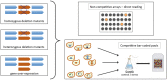
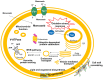
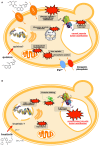
The emerging transdisciplinary field of Toxicogenomics aims to study the cell response to a given toxicant at the genome, transcriptome, proteome, and metabolome levels. This approach is expected to provide earlier and more sensitive biomarkers of toxicological responses and help in the delineation of regulatory risk assessment. The use of model organisms to gather such genomic information, through the exploitation of Omics and Bioinformatics approaches and tools, together with more focused molecular and cellular biology studies are rapidly increasing our understanding and providing an integrative view on how cells interact with their environment. The use of the model eukaryote Saccharomyces cerevisiae in the field of Toxicogenomics is discussed in this review. Despite the limitations intrinsic to the use of such a simple single cell experimental model, S. cerevisiae appears to be very useful as a first screening tool, limiting the use of animal models. Moreover, it is also one of the most interesting systems to obtain a truly global understanding of the toxicological response and resistance mechanisms, being in the frontline of systems biology research and developments. The impact of the knowledge gathered in the yeast model, through the use of Toxicogenomics approaches, is highlighted here by its use in prediction of toxicological outcomes of exposure to pesticides and pharmaceutical drugs, but also by its impact in biotechnology, namely in the development of more robust crops and in the improvement of yeast strains as cell factories.
Keywords: genome-wide approaches; molecular systems biology; predictive toxicology; response to stress; toxicity mechanisms; toxicogenomics; yeast model.
Figures

Similar articles
-
Yeast toxicogenomics: lessons from a eukaryotic cell model and cell factory.Curr Opin Biotechnol. 2015 Jun;33:183-91. doi: 10.1016/j.copbio.2015.03.001. Epub 2015 Mar 24. Curr Opin Biotechnol. 2015. PMID: 25812478 Review.
-
Toxicology and genetic toxicology in the new era of "toxicogenomics": impact of "-omics" technologies.Mutat Res. 2002 Jan 29;499(1):13-25. doi: 10.1016/s0027-5107(01)00292-5. Mutat Res. 2002. PMID: 11804602 Review.
-
Toxicogenomics concepts and applications to study hepatic effects of food additives and chemicals.Toxicol Appl Pharmacol. 2005 Sep 1;207(2 Suppl):179-88. doi: 10.1016/j.taap.2005.01.050. Toxicol Appl Pharmacol. 2005. PMID: 16139318 Review.
-
Toxicogenomics Approaches to Address Toxicity and Carcinogenicity in the Liver.Toxicol Pathol. 2023 Oct;51(7-8):470-481. doi: 10.1177/01926233241227942. Epub 2024 Jan 30. Toxicol Pathol. 2023. PMID: 38288963 Free PMC article. Review.
-
Environmental genomics: mechanistic insights into toxicity of and resistance to the herbicide 2,4-D.Trends Biotechnol. 2007 Aug;25(8):363-70. doi: 10.1016/j.tibtech.2007.06.002. Epub 2007 Jun 18. Trends Biotechnol. 2007. PMID: 17576017 Review.
Cited by
-
The effect of acetaminophen on ubiquitin homeostasis in Saccharomyces cerevisiae.PLoS One. 2017 Mar 14;12(3):e0173573. doi: 10.1371/journal.pone.0173573. eCollection 2017. PLoS One. 2017. PMID: 28291796 Free PMC article.
-
Harmful effects of metal(loid) oxide nanoparticles.Appl Microbiol Biotechnol. 2021 Feb;105(4):1379-1394. doi: 10.1007/s00253-021-11124-1. Epub 2021 Feb 1. Appl Microbiol Biotechnol. 2021. PMID: 33521847 Free PMC article. Review.
-
Proteomic analysis of the S. cerevisiae response to the anticancer ruthenium complex KP1019.Metallomics. 2020 Jun 24;12(6):876-890. doi: 10.1039/d0mt00008f. Metallomics. 2020. PMID: 32329475 Free PMC article.
-
Transcriptomic responses of the basidiomycete yeast Sporobolomyces sp. to the mycotoxin patulin.BMC Genomics. 2016 Mar 9;17:210. doi: 10.1186/s12864-016-2550-4. BMC Genomics. 2016. PMID: 26956724 Free PMC article.
-
A genome-wide screen identifies yeast genes required for tolerance to technical toxaphene, an organochlorinated pesticide mixture.PLoS One. 2013 Nov 18;8(11):e81253. doi: 10.1371/journal.pone.0081253. eCollection 2013. PLoS One. 2013. PMID: 24260565 Free PMC article.
References
-
- Abdulrehman D., Monteiro P. T., Teixeira M. C., Mira N. P., Lourenco A. B., dos Santos S. C., Cabrito T. R., Francisco A. P., Madeira S. C., Aires R. S., Oliveira A. L., Sa-Correia I., Freitas A. T. (2011). YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 39, D136–D14010.1093/nar/gkr646 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

