Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer
- PMID: 22530051
- PMCID: PMC3329481
- DOI: 10.1371/journal.pone.0035569
Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer
Abstract
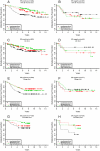
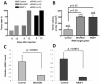
Breast cancers can recur after removal of the primary tumor and treatment to eliminate remaining tumor cells. Recurrence may occur after long periods of time during which there are no clinical symptoms. Tumor cell dormancy may explain these prolonged periods of asymptomatic residual disease and treatment resistance. We generated a dormancy gene signature from published experimental models and applied it to both breast cancer cell line expression data as well as four published clinical studies of primary breast cancers. We found that estrogen receptor (ER) positive breast cell lines and primary tumors have significantly higher dormancy signature scores (P<0.0000001) than ER- cell lines and tumors. In addition, a stratified analysis combining all ER+ tumors in four studies indicated 2.1 times higher hazard of recurrence among patients whose tumors had low dormancy scores (LDS) compared to those whose tumors had high dormancy scores (HDS) (p<0.000005). The trend was shown in all four individual studies. Suppression of two dormancy genes, BHLHE41 and NR2F1, resulted in increased in vivo growth of ER positive MCF7 cells. The patient data analysis suggests that disseminated ER positive tumor cells carrying a dormancy signature are more likely to undergo prolonged dormancy before resuming metastatic growth. Furthermore, genes identified with this approach might provide insight into the mechanisms of dormancy onset and maintenance as well as dormancy models using human breast cancer cell lines.
Conflict of interest statement
Figures

References
-
- Naumov GN, Townson JL, MacDonald IC, Wilson SM, Bramwell VH, et al. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat. 2003;82:199–206. - PubMed
-
- Muller V, Stahmann N, Riethdorf S, Rau T, Zabel T, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;11:3678–3685. - PubMed
-
- Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. - PubMed
-
- Vessella RL, Pantel K, Mohla S. Tumor Cell Dormancy: An NCI Workshop Report. Cancer Biol Ther. 2007;6 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR025750/RR/NCRR NIH HHS/United States
- 1UL1RR025750-0/RR/NCRR NIH HHS/United States
- U54 CA163131/CA/NCI NIH HHS/United States
- R01 CA109182/CA/NCI NIH HHS/United States
- CA1000324/CA/NCI NIH HHS/United States
- ES017146/ES/NIEHS NIH HHS/United States
- CA163131/CA/NCI NIH HHS/United States
- CA109182/CA/NCI NIH HHS/United States
- R21 ES017146/ES/NIEHS NIH HHS/United States
- R01 CA077522/CA/NCI NIH HHS/United States
- 5P30CA013330-37/CA/NCI NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- CA77522/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

