Gut microbiome of the critically endangered New Zealand parrot, the kakapo (Strigops habroptilus)
- PMID: 22530070
- PMCID: PMC3329475
- DOI: 10.1371/journal.pone.0035803
Gut microbiome of the critically endangered New Zealand parrot, the kakapo (Strigops habroptilus)
Abstract
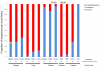
The kakapo, a parrot endemic to New Zealand, is currently the focus of intense research and conservation efforts with the aim of boosting its population above the current 'critically endangered' status. While virtually nothing is known about the microbiology of the kakapo, given the acknowledged importance of gut-associated microbes in vertebrate nutrition and pathogen defense, it should be of great conservation value to analyze the microbes associated with kakapo. Here we describe the first study of the bacterial communities that reside within the gastrointestinal tract (GIT) of both juvenile and adult kakapo. Samples from along the GIT, taken from the choana (≈ throat), crop and faeces, were subjected to 16 S rRNA gene library analysis. Phylogenetic analysis of >1000 16 S rRNA gene clones, derived from six birds, revealed low phylum-level diversity, consisting almost exclusively of Firmicutes (including lactic acid bacteria) and Gammaproteobacteria. The relative proportions of Firmicutes and Gammaproteobacteria were highly consistent among individual juveniles, irrespective of sampling location, but differed markedly among adult birds. Diversity at a finer phylogenetic resolution (i.e. operational taxonomic units (OTUs) of 99% sequence identity) was also low in all samples, with only one or two OTUs dominating each sample. These data represent the first analysis of the bacterial communities associated with the kakapo GIT, providing a baseline for further microbiological study, and facilitating conservation efforts for this unique bird.
Conflict of interest statement
Figures



References
-
- Merton DV, Morris RB, Atkinson IAE. Lek behaviour in a parrot: the Kakapo Strigops habroptilus of New Zealand. Ibis. 1984;126:277–283.
-
- Lloyd BD, Powlesland RG. The decline of kakapo Strigops habroptilus and attempts at conservation by translocation. Biol Conserv. 1994;69:75–85.
-
- Houston D, Mcinnes K, Elliott G, Eason D, Moorhouse R, et al. The use of a nutritional supplement to improve egg production in the endangered kakapo. Biol Conserv. 2007;138:248–255.
-
- Elliott GP, Merton DV, Jansen PW. Intensive management of a critically endangered species: the kakapo. Biol Conserv. 2001;99:121–133.
-
- Dubos R, Schaedler RW. The digestive tract as an ecosystem. Am J Med Sci. 1964;248:267–272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

