Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1
- PMID: 22531783
- PMCID: PMC3365429
- DOI: 10.1038/emboj.2012.92
Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1
Erratum in
-
Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1.EMBO J. 2015 Oct 14;34(20):2593-4. doi: 10.15252/embj.201570070. Epub 2015 Aug 20. EMBO J. 2015. PMID: 26294795 Free PMC article. No abstract available.
Abstract
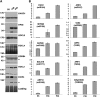
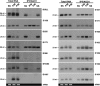
Plant viruses encode RNA silencing suppressors (VSRs) to counteract the antiviral RNA silencing response. Based on in-vitro studies, several VSRs were proposed to suppress silencing through direct binding of short-interfering RNAs (siRNAs). Because their expression also frequently hinders endogenous miRNA-mediated regulation and stabilizes labile miRNA* strands, VSRs have been assumed to prevent both siRNA and miRNA loading into their common effector protein, AGO1, through sequestration of small RNA (sRNA) duplexes in vivo. These assumptions, however, have not been formally tested experimentally. Here, we present a systematic in planta analysis comparing the effects of four distinct VSRs in Arabidopsis. While all of the VSRs tested compromised loading of siRNAs into AGO1, only P19 was found to concurrently prevent miRNA loading, consistent with a VSR strategy primarily based on sRNA sequestration. By contrast, we provide multiple lines of evidence that the action of the other VSRs tested is unlikely to entail siRNA sequestration, indicating that in-vitro binding assays and in-vivo miRNA* stabilization are not reliable indicator of VSR action. The contrasted effects of VSRs on siRNA versus miRNA loading into AGO1 also imply the existence of two distinct pools of cellular AGO1 that are specifically loaded by each class of sRNAs. These findings have important implications for our current understanding of RNA silencing and of its suppression in plants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



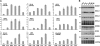
References
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 - PubMed
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
-
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC (2007) The Polerovirus silencing SUppresor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

