Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK
- PMID: 22532557
- PMCID: PMC3340917
- DOI: 10.1128/mBio.00035-12
Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK
Abstract
Although virulence of Streptococcus pneumoniae is associated with its capsule, some pathogenic S. pneumoniae isolates lack capsules and are serologically nontypeable (NT). We obtained 64 isolates that were identified as NT "pneumococci" (i.e., bacteria satisfying the conventional definition but without the multilocus sequence typing [MLST]-based definition of S. pneumoniae) by the traditional criteria. All 64 were optochin sensitive and had lytA, and 63 had ply. Twelve isolates had cpsA, suggesting the presence of a conventional but defective capsular polysaccharide synthesis (cps) locus. The 52 cpsA-negative isolates could be divided into three null capsule clades (NCC) based on aliC (aliB-like ORF1), aliD (aliB-like ORF2), and our newly discovered gene, pspK, in their cps loci. pspK encodes a protein with a long alpha-helical region containing an LPxTG motif and a YPT motif known to bind human pIgR. There were nine isolates in NCC1 (pspK(+) but negative for aliC and aliD), 32 isolates in NCC2 (aliC(+) aliD(+) but negative for pspK), and 11 in NCC3 (aliD(+) but negative for aliC and pspK). Among 52 cpsA-negative isolates, 41 were identified as S. pneumoniae by MLST analysis. All NCC1 and most NCC2 isolates were S. pneumoniae, whereas all nine NCC3 and two NCC2 isolates were not S. pneumoniae. Several NCC1 and NCC2 isolates from multiple individuals had identical MLST and cps regions, showing that unencapsulated S. pneumoniae can be infectious among humans. Furthermore, NCC1 and NCC2 S. pneumoniae isolates could colonize mice as well as encapsulated S. pneumoniae, although S. pneumoniae with an artificially disrupted cps locus did not. Moreover, an NCC1 isolate with pspK deletion did not colonize mice, suggesting that pspK is critical for colonization. Thus, PspK may provide pneumococci a means of surviving in the nasopharynx without capsule. IMPORTANCE The presence of a capsule is critical for many pathogenic bacteria, including pneumococci. Reflecting the pathogenic importance of the pneumococcal capsule, pneumococcal vaccines are designed to elicit anticapsule antibodies. Additional evidence for the pathogenic importance of the pneumococcal capsule is the fact that in pneumococci all the genes necessary for capsule production are together in one genetic locus, which is called the cps locus. However, there are occasional pathogenic pneumococci without capsules, and how they survive in the host without the capsule is unknown. Here, we show that in these acapsular pneumococci, the cps loci have been replaced with various novel genes and they can colonize mouse nasopharynges as well as capsulated pneumococci. Since the genes that replace the cps loci are likely to be important in host survival, they may show new and/or alternative capsule-independent survival mechanisms used by pneumococci.
Figures


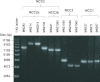
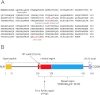

References
-
- Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
