Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal
- PMID: 22532567
- PMCID: PMC3366007
- DOI: 10.1074/jbc.M112.352930
Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal
Abstract
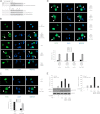
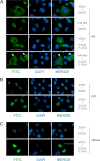
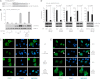
Synthesis of truncated androgen receptor (AR) splice variants has emerged as an important mechanism of prostate cancer (PCa) resistance to AR-targeted therapy and progression to a lethal castration-resistant phenotype. However, the precise role of these factors at this stage of the disease is not clear due to loss of multiple COOH-terminal AR protein domains, including the canonical nuclear localization signal (NLS) in the AR hinge region. Despite loss of this NLS, we show that diverse truncated AR variant species have a basal level of nuclear localization sufficient for ligand-independent transcriptional activity. Whereas full-length AR requires Hsp90 and importin-β for active nuclear translocation, basal nuclear localization of truncated AR variants is independent of these classical signals. For a subset of truncated AR variants, this basal level of nuclear import can be augmented by unique COOH-terminal sequences that reconstitute classical AR NLS activity. However, this property is separable from ligand-independent transcriptional activity. Therefore, the AR splice variant core consisting of the AR NH(2)-terminal domain and DNA binding domain is sufficient for nuclear localization and androgen-independent transcriptional activation of endogenous AR target genes. Indeed, we show that truncated AR variants with nuclear as well as nuclear/cytoplasmic localization patterns can drive androgen-independent growth of PCa cells. Together, our data demonstrate that diverse truncated AR species with varying efficiencies of nuclear localization can contribute to castration-resistant PCa pathology by driving persistent ligand-independent AR transcriptional activity.
Figures

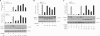



References
-
- Dehm S. M., Tindall D. J. (2007) Androgen receptor structural and functional elements. Role and regulation in prostate cancer. Mol. Endocrinol. 21, 2855–2863 - PubMed
-
- Garraway L. A., Sellers W. R. (2006) Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 6, 593–602 - PubMed
-
- de Bono J. S., Logothetis C. J., Molina A., Fizazi K., North S., Chu L., Chi K. N., Jones R. J., Goodman O. B., Jr., Saad F., Staffurth J. N., Mainwaring P., Harland S., Flaig T. W., Hutson T. E., Cheng T., Patterson H., Hainsworth J. D., Ryan C. J., Sternberg C. N., Ellard S. L., Fléchon A., Saleh M., Scholz M., Efstathiou E., Zivi A., Bianchini D., Loriot Y., Chieffo N., Kheoh T., Haqq C. M., Scher H. I., and COU-AA-301 Investigators (2011) Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

