Slow fitness recovery in a codon-modified viral genome
- PMID: 22532576
- PMCID: PMC3457771
- DOI: 10.1093/molbev/mss119
Slow fitness recovery in a codon-modified viral genome
Abstract
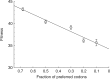
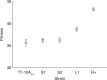
Extensive synonymous codon modification of viral genomes appears to be an effective way of attenuating strains for use as live vaccines. An assumption of this method is that codon changes have individually small effects, such that codon-attenuated viruses will be slow to evolve back to high fitness (and thus to high virulence). The major capsid gene of the bacterial virus T7 was modified to have varying levels of suboptimal synonymous codons in different constructs, and fitnesses declined linearly with the number of changes. Adaptation of the most extreme design, with 182 codon changes, resulted in a slow fitness recovery by standards of previous experimental evolution with this virus, although fitness effects of substitutions were higher than expected from the average effect of an engineered codon modification. Molecular evolution during recovery was modest, and changes evolved both within the modified gene and outside it. Some changes within the modified gene evolved in parallel across replicates, but with no obvious explanation. Overall, the study supports the premise that codon-modified viruses recover fitness slowly, although the evolution is substantially more rapid than expected from the design principle.
Figures
References
-
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

