Computational design of a PDZ domain peptide inhibitor that rescues CFTR activity
- PMID: 22532795
- PMCID: PMC3330111
- DOI: 10.1371/journal.pcbi.1002477
Computational design of a PDZ domain peptide inhibitor that rescues CFTR activity
Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is an epithelial chloride channel mutated in patients with cystic fibrosis (CF). The most prevalent CFTR mutation, ΔF508, blocks folding in the endoplasmic reticulum. Recent work has shown that some ΔF508-CFTR channel activity can be recovered by pharmaceutical modulators ("potentiators" and "correctors"), but ΔF508-CFTR can still be rapidly degraded via a lysosomal pathway involving the CFTR-associated ligand (CAL), which binds CFTR via a PDZ interaction domain. We present a study that goes from theory, to new structure-based computational design algorithms, to computational predictions, to biochemical testing and ultimately to epithelial-cell validation of novel, effective CAL PDZ inhibitors (called "stabilizers") that rescue ΔF508-CFTR activity. To design the "stabilizers", we extended our structural ensemble-based computational protein redesign algorithm K* to encompass protein-protein and protein-peptide interactions. The computational predictions achieved high accuracy: all of the top-predicted peptide inhibitors bound well to CAL. Furthermore, when compared to state-of-the-art CAL inhibitors, our design methodology achieved higher affinity and increased binding efficiency. The designed inhibitor with the highest affinity for CAL (kCAL01) binds six-fold more tightly than the previous best hexamer (iCAL35), and 170-fold more tightly than the CFTR C-terminus. We show that kCAL01 has physiological activity and can rescue chloride efflux in CF patient-derived airway epithelial cells. Since stabilizers address a different cellular CF defect from potentiators and correctors, our inhibitors provide an additional therapeutic pathway that can be used in conjunction with current methods.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
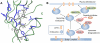

 algorithm searches over protein sequences and conformations to find the protein complexes with the best binding constant.
algorithm searches over protein sequences and conformations to find the protein complexes with the best binding constant.  takes an input model composed of an initial protein structure, a rotamer library to search over side-chain conformations, and an energy function to evaluate conformations. Minimization-aware DEE (minDEE) prunes rotamers that are not part of the lowest energy conformations for a given sequence. The remaining conformations from minDEE are enumerated in order of increasing energy lower bounds using A*. Finally, the conformations are Boltzmann-weighted and used to compute partition functions and ultimately a
takes an input model composed of an initial protein structure, a rotamer library to search over side-chain conformations, and an energy function to evaluate conformations. Minimization-aware DEE (minDEE) prunes rotamers that are not part of the lowest energy conformations for a given sequence. The remaining conformations from minDEE are enumerated in order of increasing energy lower bounds using A*. Finally, the conformations are Boltzmann-weighted and used to compute partition functions and ultimately a  score for each sequence.
score for each sequence.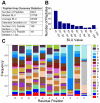
 ). (C) Normalized amino acid frequencies for the top sequences that have a BLU value greater than 3 standard deviations from the average, which were considered as the peptides that bound CAL for the validation of
). (C) Normalized amino acid frequencies for the top sequences that have a BLU value greater than 3 standard deviations from the average, which were considered as the peptides that bound CAL for the validation of  predictions. The frequency of each amino acid type for each residue position was normalized by the total number of occurrences of that amino acid in the array at the given residue position.
predictions. The frequency of each amino acid type for each residue position was normalized by the total number of occurrences of that amino acid in the array at the given residue position.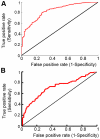
 predictions to (A) the entire HumLib peptide array data set (AUC = 0.84) and (B) only sequences in the HumLib array that matched the CAL binding motif (AUC = 0.71).
predictions to (A) the entire HumLib peptide array data set (AUC = 0.84) and (B) only sequences in the HumLib array that matched the CAL binding motif (AUC = 0.71).
 ). Horizontal line represents average
). Horizontal line represents average  G for plotted sequences. Sequence information and binding data can be found in Tables 1 and 2. (B) Ensemble of top 100 conformations for the peptide (kCAL01: WQVTRV, orange sticks) with tightest binding to CAL (gray ribbon).
G for plotted sequences. Sequence information and binding data can be found in Tables 1 and 2. (B) Ensemble of top 100 conformations for the peptide (kCAL01: WQVTRV, orange sticks) with tightest binding to CAL (gray ribbon).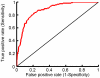
 predictions to the observed peptide array binding data. AUC = 0.88.
predictions to the observed peptide array binding data. AUC = 0.88.
 values shown are for pairwise comparisons (
values shown are for pairwise comparisons ( ). Values shown are mean
). Values shown are mean  standard error of the mean (SEM). N.S.: not significant,
standard error of the mean (SEM). N.S.: not significant,  .
.References
-
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. - PubMed
-
- Humbert P, Russell S, Richardson H. Dlg, scribble and lgl in cell polarity, cell proliferation and cancer. Bioessays. 2003;25:542–553. - PubMed
-
- Guggino WB, Stanton BA. New insights into cystic fibrosis: molecular switches that regulate CFTR. Nat Rev Mol Cell Biol. 2006;7:426–436. - PubMed
-
- Cheng J, Moyer BD, Milewski M, Loffing J, Ikeda M, et al. A golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J Biol Chem. 2002;277:3520–3529. - PubMed
-
- Cheng J, Wang H, Guggino WB. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J Biol Chem. 2004;279:1892–1898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

