Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure
- PMID: 22534784
- PMCID: PMC3352832
- DOI: 10.1093/jnci/djs191
Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure
Abstract
Background: Two single-nucleotide polymorphisms, rs1051730 and rs16969968, located within the nicotinic acetylcholine receptor gene cluster on chromosome 15q25 locus, are associated with heaviness of smoking, risk for lung cancer, and other smoking-related health outcomes. Previous studies have typically relied on self-reported smoking behavior, which may not fully capture interindividual variation in tobacco exposure.
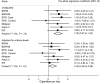
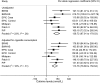
Methods: We investigated the association of rs1051730 and rs16969968 genotype (referred to as rs1051730-rs16969968, because these are in perfect linkage disequilibrium and interchangeable) with both self-reported daily cigarette consumption and biochemically measured plasma or serum cotinine levels among cigarette smokers. Summary estimates and descriptive statistical data for 12 364 subjects were obtained from six independent studies, and 2932 smokers were included in the analyses. Linear regression was used to calculate the per-allele association of rs1051730-rs16969968 genotype with cigarette consumption and cotinine levels in current smokers for each study. Meta-analysis of per-allele associations was conducted using a random effects method. The likely resulting association between genotype and lung cancer risk was assessed using published data on the association between cotinine levels and lung cancer risk. All statistical tests were two-sided.
Results: Pooled per-allele associations showed that current smokers with one or two copies of the rs1051730-rs16969968 risk allele had increased self-reported cigarette consumption (mean increase in unadjusted number of cigarettes per day per allele = 1.0 cigarette, 95% confidence interval [CI] = 0.57 to 1.43 cigarettes, P = 5.22 × 10(-6)) and cotinine levels (mean increase in unadjusted cotinine levels per allele = 138.72 nmol/L, 95% CI = 97.91 to 179.53 nmol/L, P = 2.71 × 10(-11)). The increase in cotinine levels indicated an increased risk of lung cancer with each additional copy of the rs1051730-rs16969968 risk allele (per-allele odds ratio = 1.31, 95% CI = 1.21 to 1.42).
Conclusions: Our data show a stronger association of rs1051730-rs16969968 genotype with objective measures of tobacco exposure compared with self-reported cigarette consumption. The association of these variants with lung cancer risk is likely to be mediated largely, if not wholly, via tobacco exposure.
Figures
Comment in
-
Cotinine conundrum--a step forward but questions remain.J Natl Cancer Inst. 2012 May 16;104(10):720-2. doi: 10.1093/jnci/djs211. Epub 2012 Apr 25. J Natl Cancer Inst. 2012. PMID: 22534783 Free PMC article. No abstract available.
References
-
- Hung RJ, Mckay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. - PubMed
-
- Young RP, Hophins RJ, Hay BA, Epton MJ, Black PN, Gamble GD. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur Resp J. 2008;32(5):1158–1164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

