Intuitive representation of surface properties of biomolecules using BioBlender
- PMID: 22536962
- PMCID: PMC3434447
- DOI: 10.1186/1471-2105-13-S4-S16
Intuitive representation of surface properties of biomolecules using BioBlender
Abstract
Background: In living cells, proteins are in continuous motion and interaction with the surrounding medium and/or other proteins and ligands. These interactions are mediated by protein features such as electrostatic and lipophilic potentials. The availability of protein structures enables the study of their surfaces and surface characteristics, based on atomic contribution. Traditionally, these properties are calculated by physico-chemical programs and visualized as range of colors that vary according to the tool used and imposes the necessity of a legend to decrypt it. The use of color to encode both characteristics makes the simultaneous visualization almost impossible, requiring these features to be visualized in different images. In this work, we describe a novel and intuitive code for the simultaneous visualization of these properties.
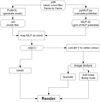
Methods: Recent advances in 3D animation and rendering software have not yet been exploited for the representation of biomolecules in an intuitive, animated form. For our purpose we use Blender, an open-source, free, cross-platform application used professionally for 3D work. On the basis Blender, we developed BioBlender, dedicated to biological work: elaboration of protein motion with simultaneous visualization of their chemical and physical features. Electrostatic and lipophilic potentials are calculated using physico-chemical software and scripts, organized and accessed through BioBlender interface.
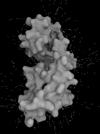
Results: A new visual code is introduced for molecular lipophilic potential: a range of optical features going from smooth-shiny for hydrophobic regions to rough-dull for hydrophilic ones. Electrostatic potential is represented as animated line particles that flow along field lines, proportional to the total charge of the protein.
Conclusions: Our system permits visualization of molecular features and, in the case of moving proteins, their continuous perception, calculated for each conformation during motion. Using real world tactile/sight feelings, the nanoscale world of proteins becomes more understandable, familiar to our everyday life, making it easier to introduce "un-seen" phenomena (concepts) such as hydropathy or charges. Moreover, this representation contributes to gain insight into molecular functions by drawing viewer's attention to the most active regions of the protein. The program, available for Windows, Linux and MacOS, can be downloaded freely from the dedicated website http://www.bioblender.eu.
Figures



References
-
- Protein Data Bank. http://www.pdb.org
-
- Visual Molecular Dynamics. http://www.ks.uiuc.edu/Research/vmd/
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

