PSI-Search: iterative HOE-reduced profile SSEARCH searching
- PMID: 22539666
- PMCID: PMC3371869
- DOI: 10.1093/bioinformatics/bts240
PSI-Search: iterative HOE-reduced profile SSEARCH searching
Abstract
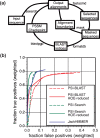
Iterative similarity searches with PSI-BLAST position-specific score matrices (PSSMs) find many more homologs than single searches, but PSSMs can be contaminated when homologous alignments are extended into unrelated protein domains-homologous over-extension (HOE). PSI-Search combines an optimal Smith-Waterman local alignment sequence search, using SSEARCH, with the PSI-BLAST profile construction strategy. An optional sequence boundary-masking procedure, which prevents alignments from being extended after they are initially included, can reduce HOE errors in the PSSM profile. Preventing HOE improves selectivity for both PSI-BLAST and PSI-Search, but PSI-Search has ~4-fold better selectivity than PSI-BLAST and similar sensitivity at 50% and 60% family coverage. PSI-Search is also produces 2- for 4-fold fewer false-positives than JackHMMER, but is ~5% less sensitive.
Availability and implementation: PSI-Search is available from the authors as a standalone implementation written in Perl for Linux-compatible platforms. It is also available through a web interface (www.ebi.ac.uk/Tools/sss/psisearch) and SOAP and REST Web Services (www.ebi.ac.uk/Tools/webservices).
Figures
References
-
- Agrawal A., Huang X. PSIBLAST_PairwiseStatSig: reordering PSI-BLAST hits using pairwise statistical significance. Bioinformatics. 2009;25:1082–1083. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

