A novel biclustering approach to association rule mining for predicting HIV-1-human protein interactions
- PMID: 22539940
- PMCID: PMC3335119
- DOI: 10.1371/journal.pone.0032289
A novel biclustering approach to association rule mining for predicting HIV-1-human protein interactions
Abstract
Identification of potential viral-host protein interactions is a vital and useful approach towards development of new drugs targeting those interactions. In recent days, computational tools are being utilized for predicting viral-host interactions. Recently a database containing records of experimentally validated interactions between a set of HIV-1 proteins and a set of human proteins has been published. The problem of predicting new interactions based on this database is usually posed as a classification problem. However, posing the problem as a classification one suffers from the lack of biologically validated negative interactions. Therefore it will be beneficial to use the existing database for predicting new viral-host interactions without the need of negative samples. Motivated by this, in this article, the HIV-1-human protein interaction database has been analyzed using association rule mining. The main objective is to identify a set of association rules both among the HIV-1 proteins and among the human proteins, and use these rules for predicting new interactions. In this regard, a novel association rule mining technique based on biclustering has been proposed for discovering frequent closed itemsets followed by the association rules from the adjacency matrix of the HIV-1-human interaction network. Novel HIV-1-human interactions have been predicted based on the discovered association rules and tested for biological significance. For validation of the predicted new interactions, gene ontology-based and pathway-based studies have been performed. These studies show that the human proteins which are predicted to interact with a particular viral protein share many common biological activities. Moreover, literature survey has been used for validation purpose to identify some predicted interactions that are already validated experimentally but not present in the database. Comparison with other prediction methods is also discussed.
Conflict of interest statement
Figures
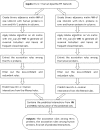

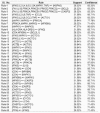
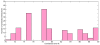

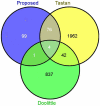
References
-
- DeFranco AL, Locksley R, Robertson M. Oxford University Press, U.K; 2007. Immunity: the immune response in infectious and inammatory disease.
-
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nature Reviews Drug Discovery. 2004;3:301–317. - PubMed
-
- Panchenko A, Przytycka T. London: Springer-Verlag; 2008. Protein-protein Interactions and Networks: Identification, Computer Analysis, and Prediction, volume 9.
-
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

