A common dataset for genomic analysis of livestock populations
- PMID: 22540034
- PMCID: PMC3337471
- DOI: 10.1534/g3.111.001453
A common dataset for genomic analysis of livestock populations
Abstract
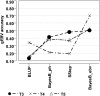
Although common datasets are an important resource for the scientific community and can be used to address important questions, genomic datasets of a meaningful size have not generally been available in livestock species. We describe a pig dataset that PIC (a Genus company) has made available for comparing genomic prediction methods. We also describe genomic evaluation of the data using methods that PIC considers best practice for predicting and validating genomic breeding values, and we discuss the impact of data structure on accuracy. The dataset contains 3534 individuals with high-density genotypes, phenotypes, and estimated breeding values for five traits. Genomic breeding values were calculated using BayesB, with phenotypes and de-regressed breeding values, and using a single-step genomic BLUP approach that combines information from genotyped and un-genotyped animals. The genomic breeding value accuracy increased with increased trait heritability and with increased relationship between training and validation. In nearly all cases, BayesB using de-regressed breeding values outperformed the other approaches, but the single-step evaluation performed only slightly worse. This dataset was useful for comparing methods for genomic prediction using real data. Our results indicate that validation approaches accounting for relatedness between populations can correct for potential overestimation of genomic breeding value accuracies, with implications for genotyping strategies to carry out genomic selection programs.
Keywords: GenPred; cross-validation; genomic relationships; pig; shared data resources.
Figures
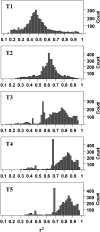
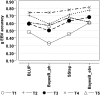

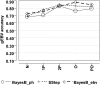
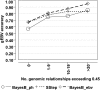
References
-
- Cleveland M. A., Forni S., Garrick D. J., Deeb N., 2010. Prediction of genomic breeding values in a commercial pig population. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production. Leipzig, Germany, August 1–6, 2010, Paper 0266
-
- Daetwyler H. D., Villanueva B., Bijma P., Woolliams J. A., 2007. Inbreeding in genome-wide selection. J. Anim. Breed. Genet. 124: 369–376 - PubMed
-
- Deeb N., Forni S., Cleveland M. A., 2010. Linkage disequilibrium decay in commercial pigs. Proceedings of the International Plant and Animal Genome XVIII Conference. San Diego, CA, January 9–13, 2010, P602
LinkOut - more resources
Full Text Sources
