Nutritional metabolomics: progress in addressing complexity in diet and health
- PMID: 22540256
- PMCID: PMC4031100
- DOI: 10.1146/annurev-nutr-072610-145159
Nutritional metabolomics: progress in addressing complexity in diet and health
Abstract
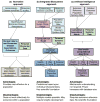
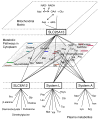
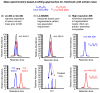
Nutritional metabolomics is rapidly maturing to use small-molecule chemical profiling to support integration of diet and nutrition in complex biosystems research. These developments are critical to facilitate transition of nutritional sciences from population-based to individual-based criteria for nutritional research, assessment, and management. This review addresses progress in making these approaches manageable for nutrition research. Important concept developments concerning the exposome, predictive health, and complex pathobiology serve to emphasize the central role of diet and nutrition in integrated biosystems models of health and disease. Improved analytic tools and databases for targeted and nontargeted metabolic profiling, along with bioinformatics, pathway mapping, and computational modeling, are now used for nutrition research on diet, metabolism, microbiome, and health associations. These new developments enable metabolome-wide association studies (MWAS) and provide a foundation for nutritional metabolomics, along with genomics, epigenomics, and health phenotyping, to support the integrated models required for personalized diet and nutrition forecasting.
Figures


References
-
- Alcock RE, Halsall CJ, Harris CA, Johnston AE, Lead WA, et al. Contamination of Environmental Samples Prepared for PCB Analysis. Environ Sci Technol. 1994;28:1838–42. - PubMed
-
- Argov Z, Chance B. Phosphorus magnetic resonance spectroscopy in nutritional research. Annu Rev Nutr. 1991;11:449–64. - PubMed
-
- Bais P, Moon SM, He K, Leitao R, Dreher K, et al. PlantMetabolomics.org: a web portal for plant metabolomics experiments. Plant Physiol. 2010;152:1807–16. - PMC - PubMed
-
- Bang JW, Crockford DJ, Holmes E, Pazos F, Sternberg MJ, et al. Integrative top-down system metabolic modeling in experimental disease states via data-driven Bayesian methods. J Proteome Res. 2008;7:497–503. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300.
Publication types
MeSH terms
Grants and funding
- R21 DK089369/DK/NIDDK NIH HHS/United States
- HL110044/HL/NHLBI NIH HHS/United States
- P50 AA013757/AA/NIAAA NIH HHS/United States
- NR012021/NR/NINR NIH HHS/United States
- ES011195/ES/NIEHS NIH HHS/United States
- P01 ES016731/ES/NIEHS NIH HHS/United States
- R01 NR012021/NR/NINR NIH HHS/United States
- DK069322/DK/NIDDK NIH HHS/United States
- R01 ES011195/ES/NIEHS NIH HHS/United States
- ES016731/ES/NIEHS NIH HHS/United States
- R21 HL110044/HL/NHLBI NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- R01 ES009047/ES/NIEHS NIH HHS/United States
- U01 DK069322/DK/NIDDK NIH HHS/United States
- UL1 RR025008/RR/NCRR NIH HHS/United States
- P20 HL113451/HL/NHLBI NIH HHS/United States
- R01 AG038746/AG/NIA NIH HHS/United States
- RR025008/RR/NCRR NIH HHS/United States
- DK089369/DK/NIDDK NIH HHS/United States
- DK089369./DK/NIDDK NIH HHS/United States
- RR023356/RR/NCRR NIH HHS/United States
- AA013757/AA/NIAAA NIH HHS/United States
- ES009047/ES/NIEHS NIH HHS/United States
- AG038746/AG/NIA NIH HHS/United States
- K24 RR023356/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources

