The transcriptional and epigenomic foundations of ground state pluripotency
- PMID: 22541430
- PMCID: PMC3398752
- DOI: 10.1016/j.cell.2012.03.026
The transcriptional and epigenomic foundations of ground state pluripotency
Abstract
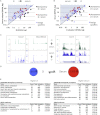
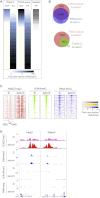
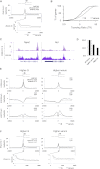
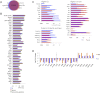
Mouse embryonic stem (ES) cells grown in serum exhibit greater heterogeneity in morphology and expression of pluripotency factors than ES cells cultured in defined medium with inhibitors of two kinases (Mek and GSK3), a condition known as "2i" postulated to establish a naive ground state. We show that the transcriptome and epigenome profiles of serum- and 2i-grown ES cells are distinct. 2i-treated cells exhibit lower expression of lineage-affiliated genes, reduced prevalence at promoters of the repressive histone modification H3K27me3, and fewer bivalent domains, which are thought to mark genes poised for either up- or downregulation. Nonetheless, serum- and 2i-grown ES cells have similar differentiation potential. Precocious transcription of developmental genes in 2i is restrained by RNA polymerase II promoter-proximal pausing. These findings suggest that transcriptional potentiation and a permissive chromatin context characterize the ground state and that exit from it may not require a metastable intermediate or multilineage priming.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures










Comment in
-
Grounded: transcriptional pausing in naive mESCs.Cell Stem Cell. 2012 May 4;10(5):484-5. doi: 10.1016/j.stem.2012.04.016. Cell Stem Cell. 2012. PMID: 22560069
References
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A.G. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q.L., Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. - PubMed
Supplemental References
-
- Ambrosino, C., Tarallo, R., Bamundo, A., Cuomo, D., Franci, G., Nassa, G., Paris, O., Ravo, M., Giovane, A., Zambrano, N., et al. (2010). Identification of a hormone-regulated dynamic nuclear actin network associated with estrogen receptor alpha in human breast cancer cell nuclei. Mol. Cell. Proteomics 9, 1352–1367. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

