Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex
- PMID: 22542188
- PMCID: PMC3340582
- DOI: 10.1016/j.neuron.2012.02.030
Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex
Abstract
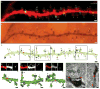
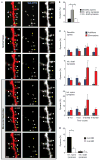
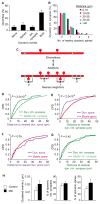
A key feature of the mammalian brain is its capacity to adapt in response to experience, in part by remodeling of synaptic connections between neurons. Excitatory synapse rearrangements have been monitored in vivo by observation of dendritic spine dynamics, but lack of a vital marker for inhibitory synapses has precluded their observation. Here, we simultaneously monitor in vivo inhibitory synapse and dendritic spine dynamics across the entire dendritic arbor of pyramidal neurons in the adult mammalian cortex using large-volume, high-resolution dual-color two-photon microscopy. We find that inhibitory synapses on dendritic shafts and spines differ in their distribution across the arbor and in their remodeling kinetics during normal and altered sensory experience. Further, we find inhibitory synapse and dendritic spine remodeling to be spatially clustered and that clustering is influenced by sensory input. Our findings provide in vivo evidence for local coordination of inhibitory and excitatory synaptic rearrangements.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Synapses let loose for a change: inhibitory synapse pruning throughout experience-dependent cortical plasticity.Neuron. 2012 Apr 26;74(2):214-7. doi: 10.1016/j.neuron.2012.04.005. Neuron. 2012. PMID: 22542175
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

