The limits to parapatric speciation: Dobzhansky-Muller incompatibilities in a continent-island model
- PMID: 22542972
- PMCID: PMC3389979
- DOI: 10.1534/genetics.111.137513
The limits to parapatric speciation: Dobzhansky-Muller incompatibilities in a continent-island model
Abstract
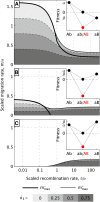
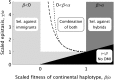
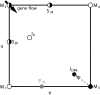
How much gene flow is needed to inhibit speciation by the accumulation of Dobzhansky-Muller incompatibilities (DMIs) in a structured population? Here, we derive these limits in a classical migration-selection model with two haploid or diploid loci and unidirectional gene flow from a continent to an island. We discuss the dependence of the maximum gene-flow rate on ecological factors (exogeneous selection), genetic factors (epistasis, recombination), and the evolutionary history. Extensive analytical and numerical results show the following: (1) The maximum rate of gene flow is limited by exogeneous selection. In particular, maintenance of neutral DMIs is impossible with gene flow. (2) There are two distinct mechanisms that drive DMI evolution in parapatry, selection against immigrants in a heterogeneous environment and selection against hybrids due to the incompatibility. (3) Depending on the mechanism, opposite predictions result concerning the genetic architecture that maximizes the rate of gene flow a DMI can sustain. Selection against immigrants favors evolution of tightly linked DMIs of arbitrary strength, whereas selection against hybrids promotes the evolution of strong unlinked DMIs. In diploids, the fitness of the double heterozygotes is the decisive factor to predict the pattern of DMI stability.
Figures


References
-
- Agrawal A. F., Feder J. L., Nosil P., 2011. Ecological divergence and the origins of intrinsic postmating isolation with gene flow. Int. J. Ecol. 2011: 1–15
-
- Bank C., Hermisson J., Kirkpatrick M., 2012. Can reinforcement complete speciation? Evolution 66: 229–239 - PubMed
-
- Barton N., Bengtsson B. O., 1986. The barrier to genetic exchange between hybridising populations. Heredity 57: 357–376 - PubMed
-
- Barton N. H., Hewitt G. M., 1989. Adaptation, speciation and hybrid zones. Nature 341: 497–503 - PubMed
-
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 Darwin and Modern Science, edited by Seward A. C. Cambridge University Press, Cambridge, UK
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

