Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells
- PMID: 22543868
- PMCID: PMC3380209
- DOI: 10.1038/emboj.2012.110
Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells
Abstract
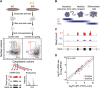
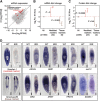
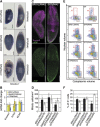
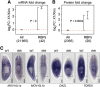
Freshwater planaria possess extreme regeneration capabilities mediated by abundant, pluripotent stem cells (neoblasts) in adult animals. Although planaria emerged as an attractive in vivo model system for stem cell biology, gene expression in neoblasts has not been profiled comprehensively and it is unknown how molecular mechanisms for pluripotency in neoblasts relate to those in mammalian embryonic stem cells (ESCs). We purified neoblasts and quantified mRNA and protein expression by sequencing and shotgun proteomics. We identified ∼4000 genes specifically expressed in neoblasts, including all ∼30 known neoblast markers. Genes important for pluripotency in ESCs, including regulators as well as targets of OCT4, were well conserved and upregulated in neoblasts. We found conserved expression of epigenetic regulators and demonstrated their requirement for planarian regeneration by knockdown experiments. Post-transcriptional regulatory genes characteristic for germ cells were also enriched in neoblasts, suggesting the existence of a common ancestral state of germ cells and ESCs. We conclude that molecular determinants of pluripotency are conserved throughout evolution and that planaria are an informative model system for human stem cell biology.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Comment in
-
A lack of commitment for over 500 million years: conserved animal stem cell pluripotency.EMBO J. 2012 Jun 13;31(12):2747-9. doi: 10.1038/emboj.2012.131. Epub 2012 May 4. EMBO J. 2012. PMID: 22562151 Free PMC article.
References
-
- Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, Dodt M, Mackowiak SD, Gogol-Doering A, Oenal P, Rybak A, Ross E, Sánchez Alvarado A, Kempa S, Dieterich C, Rajewsky N, Chen W (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res 21: 1193–1200 - PMC - PubMed
-
- Agata K (2003) Regeneration and gene regulation in planarians. Curr Opin Genet Dev 13: 492–496 - PubMed
-
- Anderson P, Kedersha N (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

