Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system: insight into intermolecular communication
- PMID: 22544739
- PMCID: PMC3375567
- DOI: 10.1074/jbc.M112.344358
Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system: insight into intermolecular communication
Abstract
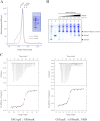
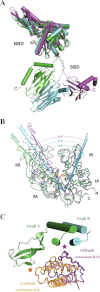
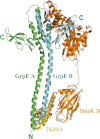
The conserved, ATP-dependent bacterial DnaK chaperones process client substrates with the aid of the co-chaperones DnaJ and GrpE. However, in the absence of structural information, how these proteins communicate with each other cannot be fully delineated. For the study reported here, we solved the crystal structure of a full-length Geobacillus kaustophilus HTA426 GrpE homodimer in complex with a nearly full-length G. kaustophilus HTA426 DnaK that contains the interdomain linker (acting as a pseudo-substrate), and the N-terminal nucleotide-binding and C-terminal substrate-binding domains at 4.1-Å resolution. Each complex contains two DnaKs and two GrpEs, which is a stoichiometry that has not been found before. The long N-terminal GrpE α-helices stabilize the linker of DnaK in the complex. Furthermore, interactions between the DnaK substrate-binding domain and the N-terminal disordered region of GrpE may accelerate substrate release from DnaK. These findings provide molecular mechanisms for substrate binding, processing, and release during the Hsp70 chaperone cycle.
Figures
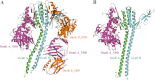
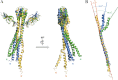


References
-
- Meimaridou E., Gooljar S. B., Chapple J. P. (2009) From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J. Mol. Endocrinol. 42, 1–9 - PubMed
-
- Hartl F. U. (1996) Molecular chaperones in cellular protein folding. Nature 381, 571–579 - PubMed
-
- Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

