The hepatitis E virus polyproline region is involved in viral adaptation
- PMID: 22545153
- PMCID: PMC3335810
- DOI: 10.1371/journal.pone.0035974
The hepatitis E virus polyproline region is involved in viral adaptation
Abstract
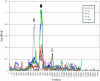
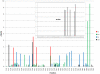
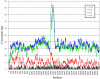
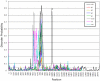
Genomes of hepatitis E virus (HEV), rubivirus and cutthroat virus (CTV) contain a region of high proline density and low amino acid (aa) complexity, named the polyproline region (PPR). In HEV genotypes 1, 3 and 4, it is the only region within the non-structural open reading frame (ORF1) with positive selection (4-10 codons with dN/dS>1). This region has the highest density of sites with homoplasy values >0.5. Genotypes 3 and 4 show ∼3-fold increase in homoplastic density (HD) in the PPR compared to any other region in ORF1, genotype 1 does not exhibit significant HD (p<0.0001). PPR sequence divergence was found to be 2-fold greater for HEV genotypes 3 and 4 than for genotype 1. The data suggest the PPR plays an important role in host-range adaptation. Although the PPR appears to be hypervariable and homoplastic, it retains as much phylogenetic signal as any other similar sized region in the ORF1, indicating that convergent evolution operates within the major HEV phylogenetic lineages. Analyses of sequence-based secondary structure and the tertiary structure identify PPR as an intrinsically disordered region (IDR), implicating its role in regulation of replication. The identified propensity for the disorder-to-order state transitions indicates the PPR is involved in protein-protein interactions. Furthermore, the PPR of all four HEV genotypes contains seven putative linear binding motifs for ligands involved in the regulation of a wide number of cellular signaling processes. Structure-based analysis of possible molecular functions of these motifs showed the PPR is prone to bind a wide variety of ligands. Collectively, these data suggest a role for the PPR in HEV adaptation. Particularly as an IDR, the PPR likely contributes to fine tuning of viral replication through protein-protein interactions and should be considered as a target for development of novel anti-viral drugs.
Conflict of interest statement
Figures



References
-
- Emerson SU, Anderson D, Arankalle A, Meng XJ, Purdy MA, et al. Hepevirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy Eigth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2004. pp. 853–857.
-
- Khuroo MS. Discovery of hepatitis E: The epidemic non-A, non-B hepatitis 30 years down the memory lane. Virus Res. 2011;161:3–14. - PubMed
-
- Teshale EH, Hu DJ, Holmberg SD. The Two Faces of Hepatitis E Virus. Clin Infect Dis. 2010;51:328–334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

