Structure of an enzyme-derived phosphoprotein recognition domain
- PMID: 22545154
- PMCID: PMC3335814
- DOI: 10.1371/journal.pone.0036014
Structure of an enzyme-derived phosphoprotein recognition domain
Abstract
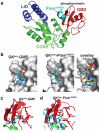
Membrane Associated Guanylate Kinases (MAGUKs) contain a protein interaction domain (GK(dom)) derived from the enzyme Guanylate Kinase (GK(enz)). Here we show that GK(dom) from the MAGUK Discs large (Dlg) is a phosphoprotein recognition domain, specifically recognizing the phosphorylated form of the mitotic spindle orientation protein Partner of Inscuteable (Pins). We determined the structure of the Dlg-Pins complex to understand the dramatic transition from nucleotide kinase to phosphoprotein recognition domain. The structure reveals that the region of the GK(dom) that once served as the GMP binding domain (GBD) has been co-opted for protein interaction. Pins makes significantly more contact with the GBD than does GMP, but primarily with residues that are conserved between enzyme and domain revealing the versatility of the GBD as a platform for nucleotide and protein interactions. Mutational analysis reveals that the GBD is also used to bind the GK ligand MAP1a, suggesting that this is a common mode of MAGUK complex assembly. The GK(enz) undergoes a dramatic closing reaction upon GMP binding but the protein-bound GK(dom) remains in the 'open' conformation indicating that the dramatic conformational change has been lost in the conversion from nucleotide kinase to phosphoprotein recognition domain.
Conflict of interest statement
Figures
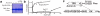


Similar articles
-
Conversion of the enzyme guanylate kinase into a mitotic-spindle orienting protein by a single mutation that inhibits GMP-induced closing.Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):E973-8. doi: 10.1073/pnas.1104365108. Epub 2011 Oct 11. Proc Natl Acad Sci U S A. 2011. PMID: 21990344 Free PMC article.
-
Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain.Biochem J. 2017 Aug 10;474(16):2817-2828. doi: 10.1042/BCJ20170356. Biochem J. 2017. PMID: 28701415
-
Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.Sci Adv. 2023 May 10;9(19):eadf3295. doi: 10.1126/sciadv.adf3295. Epub 2023 May 10. Sci Adv. 2023. PMID: 37163606 Free PMC article.
-
Mechanisms of MAGUK-mediated cellular junctional complex organization.Curr Opin Struct Biol. 2018 Feb;48:6-15. doi: 10.1016/j.sbi.2017.08.006. Epub 2017 Sep 14. Curr Opin Struct Biol. 2018. PMID: 28917202 Review.
-
Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling.Nat Rev Neurosci. 2016 Apr;17(4):209-23. doi: 10.1038/nrn.2016.18. Nat Rev Neurosci. 2016. PMID: 26988743 Review.
Cited by
-
Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.RSC Chem Biol. 2021 Jun 25;2(4):1274-1284. doi: 10.1039/d1cb00087j. eCollection 2021 Aug 5. RSC Chem Biol. 2021. PMID: 34458841 Free PMC article.
-
Metamorphic protein folding as evolutionary adaptation.Trends Biochem Sci. 2023 Aug;48(8):665-672. doi: 10.1016/j.tibs.2023.05.001. Epub 2023 Jun 1. Trends Biochem Sci. 2023. PMID: 37270322 Free PMC article. Review.
-
The Case of the Scribble Polarity Module in Asymmetric Neuroblast Division in Development and Tumorigenesis.Int J Mol Sci. 2020 Apr 20;21(8):2865. doi: 10.3390/ijms21082865. Int J Mol Sci. 2020. PMID: 32325951 Free PMC article. Review.
-
Interaction between Discs large and Pins/LGN/GPSM2: a comparison across species.Biol Open. 2021 Nov 15;10(11):bio058982. doi: 10.1242/bio.058982. Epub 2021 Nov 2. Biol Open. 2021. PMID: 34596678 Free PMC article.
-
The Drosophila mitotic spindle orientation machinery requires activation, not just localization.EMBO Rep. 2023 Mar 6;24(3):e56074. doi: 10.15252/embr.202256074. Epub 2023 Jan 11. EMBO Rep. 2023. PMID: 36629398 Free PMC article.
References
-
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. - PubMed
-
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. - PubMed
-
- Bornberg-Bauer E, Huylmans AK, Sikosek T. How do new proteins arise? Curr Opin Struct Biol. 2010;20:390–396. - PubMed
-
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

